Identification of miRNAs in extracellular vesicles as potential diagnostic markers for pediatric epilepsy and drug-resistant epilepsy via bioinformatics analysis
- PMID: 37469680
- PMCID: PMC10352456
- DOI: 10.3389/fped.2023.1199780
Identification of miRNAs in extracellular vesicles as potential diagnostic markers for pediatric epilepsy and drug-resistant epilepsy via bioinformatics analysis
Abstract
Background: Pediatric epilepsy (PE) is a common neurological disease. However, many challenges regarding the clinical diagnosis and treatment of PE and drug-resistant epilepsy (DRE) remain unsettled. Our study aimed to identify potential miRNA biomarkers in children with epilepsy and drug-resistant epilepsy by scrutinizing differential miRNA expression profiles.
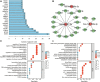
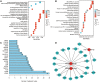
Methods: In this study, miRNA expression profiles in plasma extracellular vesicles (EV) of normal controls, children with drug-effective epilepsy (DEE), and children with DRE were obtained. In addition, differential analysis, transcription factor (TF) enrichment analysis, Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, and target gene prediction were used to identify specifically expressed miRNAs and their potential mechanisms of action. Potential diagnostic markers for DRE were identified using machine learning algorithms, and their diagnostic efficiency was assessed by the receiver operating characteristic curve (ROC).
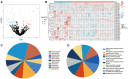
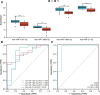
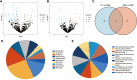
Results: The hsa-miR-1307-3p, hsa-miR-196a-5p, hsa-miR-199a-3p, and hsa-miR-21-5p were identified as diagnostic markers for PE, with values of area under curve (AUC) 0.780, 0.840, 0.832, and 0.816, respectively. In addition, the logistic regression model incorporating these four miRNAs had an AUC value of 0.940, and its target gene enrichment analysis highlighted that these miRNAs were primarily enriched in the PI3K-Akt, MAPK signaling pathways, and cell cycle. Furthermore, hsa-miR-99a-5p, hsa-miR-532-5p, hsa-miR-181d-5p, and hsa-miR-181a-5p showed good performance in differentiating children with DRE from those with DEE, with AUC values of 0.737 (0.534-0.940), 0.737 (0.523-0.952), 0.788 (0.592-0.985), and 0.788 (0.603-0.974), respectively.
Conclusion: This study characterized the expression profile of miRNAs in plasma EVs of children with epilepsy and identified miRNAs that can be used for the diagnosis of DRE.
Keywords: biomarkers; drug-resistant epilepsy; extracellular vesicles; miRNAs; pediatric epilepsy.
© 2023 Ruan, Deng, Liu, Xiao and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

