Network pharmacology and molecular docking-based analyses to predict the potential mechanism of Huangqin decoction in treating colorectal cancer
- PMID: 37469733
- PMCID: PMC10353508
- DOI: 10.12998/wjcc.v11.i19.4553
Network pharmacology and molecular docking-based analyses to predict the potential mechanism of Huangqin decoction in treating colorectal cancer
Abstract
Background: To analyze the potential action mechanism of Huangqin decoction (HQD) in colorectal cancer (CRC) treatment on the basis of network pharmacology and molecular docking.
Aim: To investigate the molecular mechanisms of HQD for CRC treatment by using network pharmacology and molecular docking.
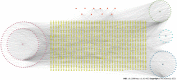
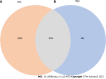
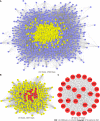
Methods: All HQD active ingredients were searched using the Systematic Pharmacology and Traditional Chinese Medicine Systems Pharmacology databases and the Bioinformatics Analysis Tool for Molecular Mechanisms in traditional Chinese medicine. Then, the targets of the active ingredients were screened. The abbreviations of protein targets were obtained from the UniProt database. A "drug-compound-target" network was constructed to screen for some main active ingredients. Some targets related to the therapeutic effect of CRC were obtained from the GeneCards, DisGeNET, Therapeutic Target Database, and Online Mendelian Inheritance in Man databases. The intersection of targets of Chinese herbs and CRC was taken. A Venn diagram was drawn to construct the intersection target interactions network by referring to the STRING database. Topological analysis of the protein interaction network was performed using Cytoscape 3.7.2 software to screen the core HQD targets for CRC. The core targets were imported into the DAVID 6.8 analysis website for gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses and visualization. Finally, molecular docking was performed using AutoDockTool and PyMOL for validation.
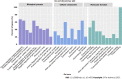
Results: In total, 280 potential drug-active ingredients were present in HQD, including 1474 targets of the drug-active ingredients. The main active ingredients identified were betulin, tetrahydropalmatine, and quercetin. In total, 10249 CRC-related targets and 1014 drug-disease intersecting targets were identified, including 28 core targets of action such as Jun proto-oncogene, AP-1 transcription factor subunit, signal transducer and activator of transcription 3, tumor protein p53, vascular endothelial growth factor, and AKT serine/threonine kinase 1. The gene ontology enrichment functional analysis yielded 503 enrichment results, including 406 biological processes that were mainly related to the positive regulation of both gene expression and transcription and cellular response to hypoxia, etc. In total, 38 cellular components were primarily related to polymer complexes, transcription factor complexes, and platelet alpha granule lumen. Then, 59 molecular functions were closely related to the binding of enzymes, homologous proteins, and transcription factors. The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis yielded 139 enrichment results, involving epidermal growth factor receptor tyrosine kinase inhibitor resistance and HIF-1 and mitogen-activated protein kinase signaling pathways.
Conclusion: HQD can play a role in CRC treatment through the "multi-component-target-pathway". The active ingredients betulin, tetrahydropalmatine, and quercetin may act on targets such as Jun proto-oncogene, AP-1 transcription factor subunit, signal transducer and activator of transcription 3, tumor protein p53, vascular endothelial growth factor, and AKT serine/threonine kinase 1, which in turn regulate HIF-1 and mitogen-activated protein kinase signaling pathways in CRC treatment. The molecular docking junction clarified that all four key target proteins could bind strongly to the main HQD active ingredients. This indicates that HQD could slow down CRC progression by modulating multiple targets and signaling pathways.
Keywords: Colorectal cancer; Huangqin decoction; Network pharmacology.
©The Author(s) 2023. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that they have no competing interests.
Figures

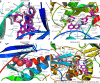


References
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. - PubMed
-
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669–685. - PubMed
-
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. - PubMed
-
- de la Chapelle A, Peltomäki P. Genetics of hereditary colon cancer. Annu Rev Genet. 1995;29:329–348. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

