The genome sequence of the King Ragworm, Alitta virens (Sars, 1835)
- PMID: 37469858
- PMCID: PMC10352625
- DOI: 10.12688/wellcomeopenres.19642.1
The genome sequence of the King Ragworm, Alitta virens (Sars, 1835)
Abstract
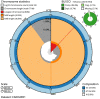
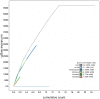
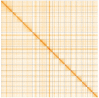
We present a genome assembly from an individual Alitta virens (the King Ragworm; Annelida; Polychaeta; Phyllodocida; Nereididae). The genome sequence is 671.2 megabases in span. Most of the assembly is scaffolded into 14 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 15.83 kilobases in length.
Keywords: Alitta virens; King Ragworm; Phyllodocida; chromosomal; genome sequence.
Copyright: © 2023 Fletcher C et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
The genome sequence of the Gelatinous Scale Worm, Alentia gelatinosa (Sars, 1835).Wellcome Open Res. 2023 Nov 22;8:542. doi: 10.12688/wellcomeopenres.20176.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38558923 Free PMC article.
-
The genome sequence of a scale worm, Harmothoe impar (Johnston, 1839).Wellcome Open Res. 2023 Jul 20;8:315. doi: 10.12688/wellcomeopenres.19570.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37786780 Free PMC article.
-
The genome sequence of the segmented worm, Sthenelais limicola (Ehlers, 1864).Wellcome Open Res. 2023 Jan 19;8:31. doi: 10.12688/wellcomeopenres.18856.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37469856 Free PMC article.
-
The genome sequence of the scale worm, Lepidonotus clava (Montagu, 1808).Wellcome Open Res. 2022 Dec 21;7:307. doi: 10.12688/wellcomeopenres.18660.1. eCollection 2022. Wellcome Open Res. 2022. PMID: 37362008 Free PMC article.
-
Revision of the Alitta virens species complex (Annelida: Nereididae) from the North Pacific Ocean.Zootaxa. 2018 Sep 21;4483(2):201-257. doi: 10.11646/zootaxa.4483.2.1. Zootaxa. 2018. PMID: 30313786
Cited by
-
Taxonomic Distribution and Molecular Evolution of Mytilectins.Mar Drugs. 2023 Nov 27;21(12):614. doi: 10.3390/md21120614. Mar Drugs. 2023. PMID: 38132935 Free PMC article. Review.
-
Annelid Comparative Genomics and the Evolution of Massive Lineage-Specific Genome Rearrangement in Bilaterians.Mol Biol Evol. 2024 Sep 4;41(9):msae172. doi: 10.1093/molbev/msae172. Mol Biol Evol. 2024. PMID: 39141777 Free PMC article.
-
Nematostella vectensis exemplifies the exceptional expansion and diversity of opsins in the eyeless Hexacorallia.Evodevo. 2023 Sep 21;14(1):14. doi: 10.1186/s13227-023-00218-8. Evodevo. 2023. PMID: 37735470 Free PMC article.
-
Acceleration of genome rearrangement in clitellate annelids.bioRxiv [Preprint]. 2024 May 14:2024.05.12.593736. doi: 10.1101/2024.05.12.593736. bioRxiv. 2024. PMID: 38798472 Free PMC article. Preprint.
References
-
- Chambers SJ, Garwood PR: Polychaetes from Scottish waters. A guide to identification. Part 3. Family Nereidae.National Museums of Scotland,1992. 10.1017/S0025315400060197 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

