Different SWI/SNF complexes coordinately promote R-loop- and RAD52-dependent transcription-coupled homologous recombination
- PMID: 37470997
- PMCID: PMC10516656
- DOI: 10.1093/nar/gkad609
Different SWI/SNF complexes coordinately promote R-loop- and RAD52-dependent transcription-coupled homologous recombination
Abstract
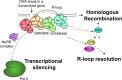
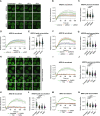
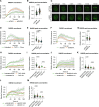
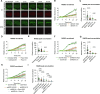
The SWI/SNF family of ATP-dependent chromatin remodeling complexes is implicated in multiple DNA damage response mechanisms and frequently mutated in cancer. The BAF, PBAF and ncBAF complexes are three major types of SWI/SNF complexes that are functionally distinguished by their exclusive subunits. Accumulating evidence suggests that double-strand breaks (DSBs) in transcriptionally active DNA are preferentially repaired by a dedicated homologous recombination pathway. We show that different BAF, PBAF and ncBAF subunits promote homologous recombination and are rapidly recruited to DSBs in a transcription-dependent manner. The PBAF and ncBAF complexes promote RNA polymerase II eviction near DNA damage to rapidly initiate transcriptional silencing, while the BAF complex helps to maintain this transcriptional silencing. Furthermore, ARID1A-containing BAF complexes promote RNaseH1 and RAD52 recruitment to facilitate R-loop resolution and DNA repair. Our results highlight how multiple SWI/SNF complexes perform different functions to enable DNA repair in the context of actively transcribed genes.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lans H., Hoeijmakers J.H.J., Vermeulen W., Marteijn J.A.. The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 2019; 20:766–784. - PubMed
-
- Jia N., Guo C., Nakazawa Y., van den Heuvel D., Luijsterburg M.S., Ogi T.. Dealing with transcription-blocking DNA damage: repair mechanisms, RNA polymerase II processing and human disorders. DNA Repair (Amst.). 2021; 106:103192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

