Poly(GR) interacts with key stress granule factors promoting its assembly into cytoplasmic inclusions
- PMID: 37471224
- PMCID: PMC10528326
- DOI: 10.1016/j.celrep.2023.112822
Poly(GR) interacts with key stress granule factors promoting its assembly into cytoplasmic inclusions
Abstract
C9orf72 repeat expansions are the most common genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). Poly(GR) proteins are toxic to neurons by forming cytoplasmic inclusions that sequester RNA-binding proteins including stress granule (SG) proteins. However, little is known of the factors governing poly(GR) inclusion formation. Here, we show that poly(GR) infiltrates a finely tuned network of protein-RNA interactions underpinning SG formation. It interacts with G3BP1, the key driver of SG assembly and a protein we found is critical for poly(GR) inclusion formation. Moreover, we discovered that N6-methyladenosine (m6A)-modified mRNAs and m6A-binding YTHDF proteins not only co-localize with poly(GR) inclusions in brains of c9FTD/ALS mouse models and patients with c9FTD, they promote poly(GR) inclusion formation via the incorporation of RNA into the inclusions. Our findings thus suggest that interrupting interactions between poly(GR) and G3BP1 or YTHDF1 proteins or decreasing poly(GR) altogether represent promising therapeutic strategies to combat c9FTD/ALS pathogenesis.
Keywords: CP: Molecular biology; CP: Neuroscience; G3BP1/2; YTHDF proteins; amyotrophic lateral sclerosis; chromosome 9 open reading frame 72; cytoplasmic poly(GR) inclusions; dipeptide repeat proteins; frontotemporal dementia; liquid-liquid phase separation; m6A-modified RNAs.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
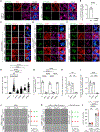


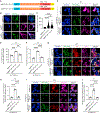
References
-
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. (2014). Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225. 10.1016/j.neuron.2014.12.010. - DOI - PMC - PubMed
-
- Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, et al. (2018). Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142. 10.1038/s41591-018-0071-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 NS097273/NS/NINDS NIH HHS/United States
- RF1 AG062171/AG/NIA NIH HHS/United States
- U01 AG072577/AG/NIA NIH HHS/United States
- RF1 AG062077/AG/NIA NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- R21 NS127331/NS/NINDS NIH HHS/United States
- R01 NS117461/NS/NINDS NIH HHS/United States
- RF1 AG061706/AG/NIA NIH HHS/United States
- P01 NS084974/NS/NINDS NIH HHS/United States
- R01 AG080810/AG/NIA NIH HHS/United States
- P01 NS099114/NS/NINDS NIH HHS/United States
- U54 NS123743/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

