Cancer-Associated Fibroblast-Like Tumor Cells Remodel the Ewing Sarcoma Tumor Microenvironment
- PMID: 37471463
- PMCID: PMC10801911
- DOI: 10.1158/1078-0432.CCR-23-1111
Cancer-Associated Fibroblast-Like Tumor Cells Remodel the Ewing Sarcoma Tumor Microenvironment
Abstract
Purpose: Despite limited genetic and histologic heterogeneity, Ewing sarcoma (EwS) tumor cells are transcriptionally heterogeneous and display varying degrees of mesenchymal lineage specification in vitro. In this study, we investigated if and how transcriptional heterogeneity of EwS cells contributes to heterogeneity of tumor phenotypes in vivo.
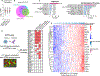
Experimental design: Single-cell proteogenomic-sequencing of EwS cell lines was performed and integrated with patient tumor transcriptomic data. Cell subpopulations were isolated by FACS for assessment of gene expression and phenotype. Digital spatial profiling and human whole transcriptome analysis interrogated transcriptomic heterogeneity in EwS xenografts. Tumor cell subpopulations and matrix protein deposition were evaluated in xenografts and patient tumors using multiplex immunofluorescence staining.
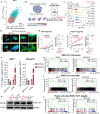
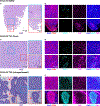
Results: We identified CD73 as a biomarker of highly mesenchymal EwS cell subpopulations in tumor models and patient biopsies. CD73+ tumor cells displayed distinct transcriptional and phenotypic properties, including selective upregulation of genes that are repressed by EWS::FLI1, and increased migratory potential. CD73+ cells were distinguished in vitro and in vivo by increased expression of matrisomal genes and abundant deposition of extracellular matrix (ECM) proteins. In epithelial-derived malignancies, ECM is largely deposited by cancer-associated fibroblasts (CAF), and we thus labeled CD73+ EwS cells, CAF-like tumor cells. Marked heterogeneity of CD73+ EwS cell frequency and distribution was detected in tumors in situ, and CAF-like tumor cells and associated ECM were observed in peri-necrotic regions and invasive foci.
Conclusions: EwS tumor cells can adopt CAF-like properties, and these distinct cell subpopulations contribute to tumor heterogeneity by remodeling the tumor microenvironment. See related commentary by Kuo and Amatruda, p. 5002.
©2023 American Association for Cancer Research.
Conflict of interest statement
Figures



Update of
-
Carcinoma-associated fibroblast-like tumor cells remodel the Ewing sarcoma tumor microenvironment.bioRxiv [Preprint]. 2023 Apr 13:2023.04.12.536619. doi: 10.1101/2023.04.12.536619. bioRxiv. 2023. Update in: Clin Cancer Res. 2023 Dec 15;29(24):5140-5154. doi: 10.1158/1078-0432.CCR-23-1111. PMID: 37090655 Free PMC article. Updated. Preprint.
Comment in
-
Seed Becoming Soil: A New Paradigm of the Ewing Sarcoma Tumor Microenvironment.Clin Cancer Res. 2023 Dec 15;29(24):5002-5004. doi: 10.1158/1078-0432.CCR-23-2259. Clin Cancer Res. 2023. PMID: 37796143 Free PMC article.
References
-
- McGranahan N, and Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–28. - PubMed
-
- Hanahan D Hallmarks of Cancer: New Dimensions. Cancer Discovery. 2022;12(1):31–46. - PubMed
-
- Li Z, Seehawer M, and Polyak K. Untangling the web of intratumour heterogeneity. Nature cell biology. 2022;24(8):1192–201. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

