Risk averse reproduction numbers improve resurgence detection
- PMID: 37471464
- PMCID: PMC10393178
- DOI: 10.1371/journal.pcbi.1011332
Risk averse reproduction numbers improve resurgence detection
Abstract
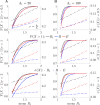
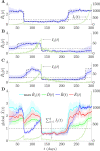
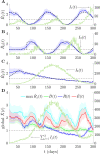
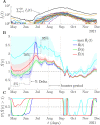
The effective reproduction number R is a prominent statistic for inferring the transmissibility of infectious diseases and effectiveness of interventions. R purportedly provides an easy-to-interpret threshold for deducing whether an epidemic will grow (R>1) or decline (R<1). We posit that this interpretation can be misleading and statistically overconfident when applied to infections accumulated from groups featuring heterogeneous dynamics. These groups may be delineated by geography, infectiousness or sociodemographic factors. In these settings, R implicitly weights the dynamics of the groups by their number of circulating infections. We find that this weighting can cause delayed detection of outbreak resurgence and premature signalling of epidemic control because it underrepresents the risks from highly transmissible groups. Applying E-optimal experimental design theory, we develop a weighting algorithm to minimise these issues, yielding the risk averse reproduction number E. Using simulations, analytic approaches and real-world COVID-19 data stratified at the city and district level, we show that E meaningfully summarises transmission dynamics across groups, balancing bias from the averaging underlying R with variance from directly using local group estimates. An E>1generates timely resurgence signals (upweighting risky groups), while an E<1ensures local outbreaks are under control. We propose E as an alternative to R for informing policy and assessing transmissibility at large scales (e.g., state-wide or nationally), where R is commonly computed but well-mixed or homogeneity assumptions break down.
Copyright: © 2023 Parag, Obolski. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
We declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

