Mitochondrial double-stranded RNA triggers induction of the antiviral DNA deaminase APOBEC3A and nuclear DNA damage
- PMID: 37474103
- PMCID: PMC10457583
- DOI: 10.1016/j.jbc.2023.105073
Mitochondrial double-stranded RNA triggers induction of the antiviral DNA deaminase APOBEC3A and nuclear DNA damage
Abstract
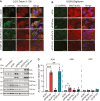
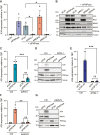
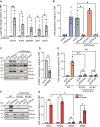
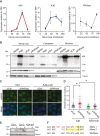
APOBEC3A is an antiviral DNA deaminase often induced by virus infection. APOBEC3A is also a source of cancer mutation in viral and nonviral tumor types. It is therefore critical to identify factors responsible for APOBEC3A upregulation. Here, we test the hypothesis that leaked mitochondrial (mt) double-stranded (ds)RNA is recognized as foreign nucleic acid, which triggers innate immune signaling, APOBEC3A upregulation, and DNA damage. Knockdown of an enzyme responsible for degrading mtdsRNA, the exoribonuclease polynucleotide phosphorylase, results in mtdsRNA leakage into the cytosol and induction of APOBEC3A expression. APOBEC3A upregulation by cytoplasmic mtdsRNA requires RIG-I, MAVS, and STAT2 and is likely part of a broader type I interferon response. Importantly, although mtdsRNA-induced APOBEC3A appears cytoplasmic by subcellular fractionation experiments, its induction triggers an overt DNA damage response characterized by elevated nuclear γ-H2AX staining. Thus, mtdsRNA dysregulation may induce APOBEC3A and contribute to observed genomic instability and mutation signatures in cancer.
Keywords: APOBEC3A; DNA damage response; cancer mutagenesis; innate immune signaling; mitochondrial dsRNA.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
References
-
- Cervantes-Gracia K., Gramalla-Schmitz A., Weischedel J., Chahwan R. APOBECs orchestrate genomic and epigenomic editing across health and disease. Trends Genet. 2021;37:1028–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

