The SUMO-NIP45 pathway processes toxic DNA catenanes to prevent mitotic failure
- PMID: 37474739
- PMCID: PMC10497417
- DOI: 10.1038/s41594-023-01045-0
The SUMO-NIP45 pathway processes toxic DNA catenanes to prevent mitotic failure
Abstract
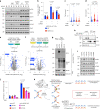
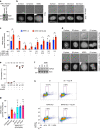
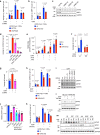
SUMOylation regulates numerous cellular processes, but what represents the essential functions of this protein modification remains unclear. To address this, we performed genome-scale CRISPR-Cas9-based screens, revealing that the BLM-TOP3A-RMI1-RMI2 (BTRR)-PICH pathway, which resolves ultrafine anaphase DNA bridges (UFBs) arising from catenated DNA structures, and the poorly characterized protein NIP45/NFATC2IP become indispensable for cell proliferation when SUMOylation is inhibited. We demonstrate that NIP45 and SUMOylation orchestrate an interphase pathway for converting DNA catenanes into double-strand breaks (DSBs) that activate the G2 DNA-damage checkpoint, thereby preventing cytokinesis failure and binucleation when BTRR-PICH-dependent UFB resolution is defective. NIP45 mediates this new TOP2-independent DNA catenane resolution process via its SUMO-like domains, promoting SUMOylation of specific factors including the SLX4 multi-nuclease complex, which contributes to catenane conversion into DSBs. Our findings establish that SUMOylation exerts its essential role in cell proliferation by enabling resolution of toxic DNA catenanes via nonepistatic NIP45- and BTRR-PICH-dependent pathways to prevent mitotic failure.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

