Sensitive multicolor indicators for monitoring norepinephrine in vivo
- PMID: 37474807
- PMCID: PMC7615053
- DOI: 10.1038/s41592-023-01959-z
Sensitive multicolor indicators for monitoring norepinephrine in vivo
Abstract
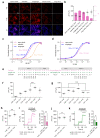
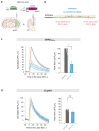
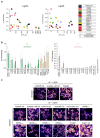
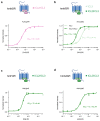
Genetically encoded indicators engineered from G-protein-coupled receptors are important tools that enable high-resolution in vivo neuromodulator imaging. Here, we introduce a family of sensitive multicolor norepinephrine (NE) indicators, which includes nLightG (green) and nLightR (red). These tools report endogenous NE release in vitro, ex vivo and in vivo with improved sensitivity, ligand selectivity and kinetics, as well as a distinct pharmacological profile compared with previous state-of-the-art GRABNE indicators. Using in vivo multisite fiber photometry recordings of nLightG, we could simultaneously monitor optogenetically evoked NE release in the mouse locus coeruleus and hippocampus. Two-photon imaging of nLightG revealed locomotion and reward-related NE transients in the dorsal CA1 area of the hippocampus. Thus, the sensitive NE indicators introduced here represent an important addition to the current repertoire of indicators and provide the means for a thorough investigation of the NE system.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
T.P. is a co-inventor on a patent application related to the technology described in this article. All other authors have nothing to disclose.
Figures











References
-
- Sara SJ. The locus coeruleus and noradrenergic modulation of cognition. Nat Rev Neurosci. 2009;10:211–223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

