Long-read, chromosome-scale assembly of Vitis rotundifolia cv. Carlos and its unique resistance to Xylella fastidiosa subsp. fastidiosa
- PMID: 37474911
- PMCID: PMC10357881
- DOI: 10.1186/s12864-023-09514-y
Long-read, chromosome-scale assembly of Vitis rotundifolia cv. Carlos and its unique resistance to Xylella fastidiosa subsp. fastidiosa
Abstract
Background: Muscadine grape (Vitis rotundifolia) is resistant to many of the pathogens that negatively impact the production of common grape (V. vinifera), including the bacterial pathogen Xylella fastidiosa subsp. fastidiosa (Xfsf), which causes Pierce's Disease (PD). Previous studies in common grape have indicated Xfsf delays host immune response with a complex O-chain antigen produced by the wzy gene. Muscadine cultivars range from tolerant to completely resistant to Xfsf, but the mechanism is unknown.
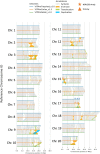
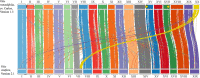
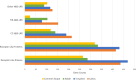
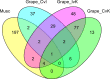
Results: We assembled and annotated a new, long-read genome assembly for 'Carlos', a cultivar of muscadine that exhibits tolerance, to build upon the existing genetic resources available for muscadine. We used these resources to construct an initial pan-genome for three cultivars of muscadine and one cultivar of common grape. This pan-genome contains a total of 34,970 synteny-constrained entries containing genes of similar structure. Comparison of resistance gene content between the 'Carlos' and common grape genomes indicates an expansion of resistance (R) genes in 'Carlos.' We further identified genes involved in Xfsf response by transcriptome sequencing 'Carlos' plants inoculated with Xfsf. We observed 234 differentially expressed genes with functions related to lipid catabolism, oxidation-reduction signaling, and abscisic acid (ABA) signaling as well as seven R genes. Leveraging public data from previous experiments of common grape inoculated with Xfsf, we determined that most differentially expressed genes in the muscadine response were not found in common grape, and three of the R genes identified as differentially expressed in muscadine do not have an ortholog in the common grape genome.
Conclusions: Our results support the utility of a pan-genome approach to identify candidate genes for traits of interest, particularly disease resistance to Xfsf, within and between muscadine and common grape.
Keywords: Grape; Muscadine grape; Pangenome; Pierce’s disease; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Proteomics approach to identify unique xylem sap proteins in Pierce's disease-tolerant Vitis species.Appl Biochem Biotechnol. 2010 Mar;160(3):932-44. doi: 10.1007/s12010-009-8620-1. Epub 2009 May 2. Appl Biochem Biotechnol. 2010. PMID: 19412582
-
Virulence Comparison of a Comprehensive Panel of Xylella fastidiosa Pierce's Disease Isolates from California.Plant Dis. 2024 Jun;108(6):1555-1564. doi: 10.1094/PDIS-09-23-1923-RE. Epub 2024 May 16. Plant Dis. 2024. PMID: 38105458
-
Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce's disease of grapevine in the U.S.PLoS One. 2010 Nov 16;5(11):e15488. doi: 10.1371/journal.pone.0015488. PLoS One. 2010. PMID: 21103383 Free PMC article.
-
Paradigms: examples from the bacterium Xylella fastidiosa.Annu Rev Phytopathol. 2013;51:339-56. doi: 10.1146/annurev-phyto-082712-102325. Epub 2013 May 17. Annu Rev Phytopathol. 2013. PMID: 23682911 Review.
-
The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology.Annu Rev Entomol. 2004;49:243-70. doi: 10.1146/annurev.ento.49.061802.123403. Annu Rev Entomol. 2004. PMID: 14651464 Review.
Cited by
-
A haplotype-resolved reference genome of Quercus alba sheds light on the evolutionary history of oaks.New Phytol. 2025 Apr;246(1):331-348. doi: 10.1111/nph.20463. Epub 2025 Feb 11. New Phytol. 2025. PMID: 39931867 Free PMC article.
-
Vitis rotundifolia Genes Introgressed with RUN1 and RPV1: Poor Recombination and Impact on V. vinifera Berry Transcriptome.Plants (Basel). 2024 Jul 29;13(15):2095. doi: 10.3390/plants13152095. Plants (Basel). 2024. PMID: 39124212 Free PMC article.
-
Exploring Pan-Genomes: An Overview of Resources and Tools for Unraveling Structure, Function, and Evolution of Crop Genes and Genomes.Biomolecules. 2023 Sep 17;13(9):1403. doi: 10.3390/biom13091403. Biomolecules. 2023. PMID: 37759803 Free PMC article. Review.
-
Rpv2 is part of a cluster of NLRs specific to Vitis rotundifolia and confers total resistance to grapevine downy mildew.Theor Appl Genet. 2025 Jul 8;138(8):177. doi: 10.1007/s00122-025-04959-z. Theor Appl Genet. 2025. PMID: 40627157
References
-
- Hoffmann M. Muscadine grape production guide for the Southeast. 2020.
-
- Hopkins DL, Mollenhauer HH, Mortensen JA. Tolerance to Pierce’s Disease and the Associated Rickettsia-like Bacterium in Muscadine Grape1. J Am Soc Hortic Sci. 1974;99:436–9. doi: 10.21273/JASHS.99.5.436. - DOI
-
- Wan Y, Schwaninger HR, Baldo AM, Labate JA, Zhong G-Y, Simon CJ. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evol Biol. 2013;13:141. doi: 10.1186/1471-2148-13-141. - DOI - PMC - PubMed
-
- Reimer FC, Hume HH, Michels J, Smith RI, Curtis RS. Scuppernong and other Muscadine grapes: origin and importance. North Carolina Agricultural Experiment Station of the College of Agriculture and Mechanic Arts; 1906.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

