Evolutionary barriers to horizontal gene transfer in macrophage-associated Salmonella
- PMID: 37475746
- PMCID: PMC10355182
- DOI: 10.1093/evlett/qrad020
Evolutionary barriers to horizontal gene transfer in macrophage-associated Salmonella
Abstract
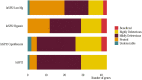
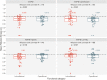
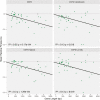
Horizontal gene transfer (HGT) is a powerful evolutionary force facilitating bacterial adaptation and emergence of novel phenotypes. Several factors, including environmental ones, are predicted to restrict HGT, but we lack systematic and experimental data supporting these predictions. Here, we address this gap by measuring the relative fitness of 44 genes horizontally transferred from Escherichia coli to Salmonella enterica in infection-relevant environments. We estimated the distribution of fitness effects in each environment and identified that dosage-dependent effects across different environments are a significant barrier to HGT. The majority of genes were found to be deleterious. We also found longer genes had stronger negative fitness consequences than shorter ones, showing that gene length was negatively associated with HGT. Furthermore, fitness effects of transferred genes were found to be environmentally dependent. In summary, a substantial fraction of transferred genes had a significant fitness cost on the recipient, with both gene characteristics and the environment acting as evolutionary barriers to HGT.
Keywords: adaptation; fitness; gene expression; microbes.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Society for the Study of Evolution (SSE) and European Society for Evolutionary Biology (ESEN).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., Huynh, W., Nguyen, A.-L. V, Cheng, A. A., Liu, S., Min, S. Y., Miroshnichenko, A., Tran, H.-K., Werfalli, R. E., Nasir, J. A., Oloni, M., Speicher, D. J., Florescu, A., Singh, B., … McArthur, A. G. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research, 48(D1), D517–D525. 10.1093/nar/gkz935 - DOI - PMC - PubMed
-
- Andrade, J. M., & Gómez-Carracedo, M. P. (2013). Notes on the use of Mandel’s test to check for nonlinearity in laboratory calibrations. Analytical Methods, 5(5), 1145–1149. 10.1039/c2ay26400e - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

