Comparative transcriptomics reveals the role of altered energy metabolism in the establishment of single-cell C4 photosynthesis in Bienertia sinuspersici
- PMID: 37476170
- PMCID: PMC10354284
- DOI: 10.3389/fpls.2023.1202521
Comparative transcriptomics reveals the role of altered energy metabolism in the establishment of single-cell C4 photosynthesis in Bienertia sinuspersici
Abstract
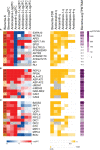
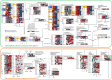
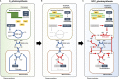
Single-cell C4 photosynthesis (SCC4) in terrestrial plants without Kranz anatomy involves three steps: initial CO2 fixation in the cytosol, CO2 release in mitochondria, and a second CO2 fixation in central chloroplasts. Here, we investigated how the large number of mechanisms underlying these processes, which occur in three different compartments, are orchestrated in a coordinated manner to establish the C4 pathway in Bienertia sinuspersici, a SCC4 plant. Leaves were subjected to transcriptome analysis at three different developmental stages. Functional enrichment analysis revealed that SCC4 cycle genes are coexpressed with genes regulating cyclic electron flow and amino/organic acid metabolism, two key processes required for the production of energy molecules in C3 plants. Comparative gene expression profiling of B. sinuspersici and three other species (Suaeda aralocaspica, Amaranthus hypochondriacus, and Arabidopsis thaliana) showed that the direction of metabolic flux was determined via an alteration in energy supply in peripheral chloroplasts and mitochondria via regulation of gene expression in the direction of the C4 cycle. Based on these results, we propose that the redox homeostasis of energy molecules via energy metabolism regulation is key to the establishment of the SCC4 pathway in B. sinuspersici.
Keywords: Amaranthus hypochondriacus; Bienertia sinuspersici; Suaeda aralocaspica; dimorphic chloroplast; malate valve; mitochondria; single-cell C4 photosynthesis; transcriptome.
Copyright © 2023 Han, Kim, Kim and Hwang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Comparative transcriptome analysis of emerging young and mature leaves of Bienertia sinuspersici, a single-cell C4 plant.PeerJ. 2025 Apr 29;13:e19282. doi: 10.7717/peerj.19282. eCollection 2025. PeerJ. 2025. PMID: 40321813 Free PMC article.
-
In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici-a comparative study across C3, C4, CAM, and SCC4 model plants.PeerJ. 2023 Jul 11;11:e15696. doi: 10.7717/peerj.15696. eCollection 2023. PeerJ. 2023. PMID: 37456874 Free PMC article.
-
The unique structural and biochemical development of single cell C4 photosynthesis along longitudinal leaf gradients in Bienertia sinuspersici and Suaeda aralocaspica (Chenopodiaceae).J Exp Bot. 2016 Apr;67(9):2587-601. doi: 10.1093/jxb/erw082. Epub 2016 Mar 8. J Exp Bot. 2016. PMID: 26957565 Free PMC article.
-
One decade after the discovery of single-cell C4 species in terrestrial plants: what did we learn about the minimal requirements of C4 photosynthesis?Photosynth Res. 2014 Feb;119(1-2):169-80. doi: 10.1007/s11120-013-9810-9. Epub 2013 Mar 14. Photosynth Res. 2014. PMID: 23494362 Review.
-
A scheme for C4 evolution derived from a comparative analysis of the closely related C3, C3-C4 intermediate, C4-like, and C4 species in the genus Flaveria.Plant Mol Biol. 2022 Nov;110(4-5):445-454. doi: 10.1007/s11103-022-01246-z. Epub 2022 Feb 4. Plant Mol Biol. 2022. PMID: 35119574 Review.
Cited by
-
Comparative transcriptome analysis of emerging young and mature leaves of Bienertia sinuspersici, a single-cell C4 plant.PeerJ. 2025 Apr 29;13:e19282. doi: 10.7717/peerj.19282. eCollection 2025. PeerJ. 2025. PMID: 40321813 Free PMC article.
-
NAD(H) and NADP(H) in plants and mammals.Mol Plant. 2025 Jun 2;18(6):938-959. doi: 10.1016/j.molp.2025.05.004. Epub 2025 May 13. Mol Plant. 2025. PMID: 40369879 Free PMC article. Review.
-
Genome-wide identification of a novel Na+ transporter from Bienertia sinuspersici and overexpression of BsHKT1;2 improved salt tolerance in Brassica rapa.Front Plant Sci. 2023 Dec 12;14:1302315. doi: 10.3389/fpls.2023.1302315. eCollection 2023. Front Plant Sci. 2023. PMID: 38192689 Free PMC article.
-
Elucidating the callus-to-shoot-forming mechanism in Capsicum annuum 'Dempsey' through comparative transcriptome analyses.BMC Plant Biol. 2024 May 7;24(1):367. doi: 10.1186/s12870-024-05033-4. BMC Plant Biol. 2024. PMID: 38711041 Free PMC article.
References
-
- Andrews S. (2010). “FastQC: a quality control tool for high throughput sequence data,” in Babraham bioinformatics (United Kingdom: Babraham Institute, Cambridge; ). Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

