Genome-wide analysis of genetic diversity in a germplasm collection including wild relatives and interspecific clones of garden asparagus
- PMID: 37476175
- PMCID: PMC10354869
- DOI: 10.3389/fpls.2023.1187663
Genome-wide analysis of genetic diversity in a germplasm collection including wild relatives and interspecific clones of garden asparagus
Abstract
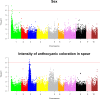
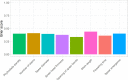
The Asparagus genus includes approximately 240 species, the most important of which is garden asparagus (Asparagus officinalis L.), as this is a vegetable crop cultivated worldwide for its edible spear. Along with garden asparagus, other species are also cultivated (e.g., Asparagus maritimus L.) or have been proposed as untapped sources of variability in breeding programs (e.g., Asparagus acutifolius L.). In the present work, we applied reduced-representation sequencing to examine a panel of 378 diverse asparagus genotypes, including commercial hybrids, interspecific lines, wild relatives of garden asparagus, and doubled haploids currently used in breeding programs, which enabled the identification of more than 200K single-nucleotide polymorphisms (SNPs). These SNPs were used to assess the extent of linkage disequilibrium in the diploid gene pool of asparagus and combined with preliminary phenotypic information to conduct genome-wide association studies for sex and traits tied to spear quality and production. Moreover, using the same phenotypic and genotypic information, we fitted and cross-validated genome-enabled prediction models for the same set of traits. Overall, our analyses demonstrated that, unlike the diversity detected in wild species related to garden asparagus and in interspecific crosses, cultivated and wild genotypes of A. officinalis L. show a narrow genetic basis, which is a contributing factor hampering the genetic improvement of this crop. Estimating the extent of linkage disequilibrium and providing the first example of genome-wide association study and genome-enabled prediction in this species, we concluded that the asparagus panel examined in the present study can lay the foundation for determination of the genetic bases of agronomically important traits and for the implementation of predictive breeding tools to sustain breeding.
Keywords: GWAS; asparagus; breeding; genetic diversity; genomic prediction; germplasm collection.
Copyright © 2023 Sala, Puglisi, Ferrari, Salamone, Tassone, Rotino, Fricano and Losa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Amato L. D., López-Anido F. S., Zayas A., Martin E. A. (2021). Genetic resources in asparagus: diversity and relationships in a collection from different origins and breeding status. New Z. J. Crop Hortic. Sci. 0, 1–12. doi: 10.1080/01140671.2021.1940217 - DOI
-
- Anido F. L., Cointry E. (2008). “Asparagus BT - vegetables II: fabaceae, liliaceae, solanaceae, and umbelliferae,” Handbook of Plant Breeding, vol 2. Eds. Prohens J., Nuez F. (New York, NY: Springer New York; ), 87–119. doi: 10.1007/978-0-387-74110-9_3 - DOI
-
- Anido F. L., Cointry E., Picardi L., Camadro E. (1997). Genetic variability of productive and vegetative characters in asparagus officinalis l.-estimates of heritability and genetic correlations. Braz. J. Genet. 20, 275–282.
LinkOut - more resources
Full Text Sources

