PCDH7 as the key gene related to the co-occurrence of sarcopenia and osteoporosis
- PMID: 37476411
- PMCID: PMC10354703
- DOI: 10.3389/fgene.2023.1163162
PCDH7 as the key gene related to the co-occurrence of sarcopenia and osteoporosis
Abstract
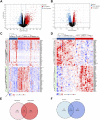
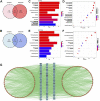
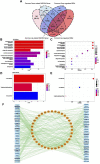
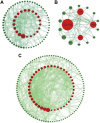
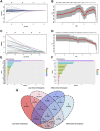
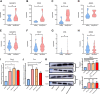
Sarcopenia and osteoporosis, two degenerative diseases in older patients, have become severe health problems in aging societies. Muscles and bones, the most important components of the motor system, are derived from mesodermal and ectodermal mesenchymal stem cells. The adjacent anatomical relationship between them provides the basic conditions for mechanical and chemical signals, which may contribute to the co-occurrence of sarcopenia and osteoporosis. Identifying the potential common crosstalk genes between them may provide new insights for preventing and treating their development. In this study, DEG analysis, WGCNA, and machine learning algorithms were used to identify the key crosstalk genes of sarcopenia and osteoporosis; this was then validated using independent datasets and clinical samples. Finally, four crosstalk genes (ARHGEF10, PCDH7, CST6, and ROBO3) were identified, and mRNA expression and protein levels of PCDH7 in clinical samples from patients with sarcopenia, with osteoporosis, and with both sarcopenia and osteoporosis were found to be significantly higher than those from patients without sarcopenia or osteoporosis. PCDH7 seems to be a key gene related to the development of both sarcopenia and osteoporosis.
Keywords: bioinformatic analysis; crosstalk genes; machine learning; osteoporosis; sarcopenia.
Copyright © 2023 Liu, Wang, Shi, Yang, Wang, Chen, Li, Chen and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification of shared gene signatures and pathways for diagnosing osteoporosis with sarcopenia through integrated bioinformatics analysis and machine learning.BMC Musculoskelet Disord. 2024 Jun 3;25(1):435. doi: 10.1186/s12891-024-07555-2. BMC Musculoskelet Disord. 2024. PMID: 38831425 Free PMC article.
-
Integrating transcriptomic and proteomic data for a comprehensive molecular perspective on the association between sarcopenia and osteoporosis.Arch Gerontol Geriatr. 2024 Oct;125:105486. doi: 10.1016/j.archger.2024.105486. Epub 2024 May 11. Arch Gerontol Geriatr. 2024. PMID: 38761527
-
Differential gene expression profile by RNA sequencing study of elderly osteoporotic hip fracture patients with sarcopenia.J Orthop Translat. 2021 May 14;29:10-18. doi: 10.1016/j.jot.2021.04.009. eCollection 2021 Jul. J Orthop Translat. 2021. PMID: 34036042 Free PMC article.
-
Sarcopenia, osteoporosis and frailty.Metabolism. 2023 Aug;145:155638. doi: 10.1016/j.metabol.2023.155638. Epub 2023 Jun 20. Metabolism. 2023. PMID: 37348597 Review.
-
Evidence for pleiotropic factors in genetics of the musculoskeletal system.Bone. 2010 May;46(5):1226-37. doi: 10.1016/j.bone.2010.01.382. Epub 2010 Feb 10. Bone. 2010. PMID: 20149904 Free PMC article. Review.
Cited by
-
Unraveling the intricacies of osteoclast differentiation and maturation: insight into novel therapeutic strategies for bone-destructive diseases.Exp Mol Med. 2024 Feb;56(2):264-272. doi: 10.1038/s12276-024-01157-7. Epub 2024 Feb 1. Exp Mol Med. 2024. PMID: 38297158 Free PMC article. Review.
-
Role and Validation of Lactylation-Related Gene Markers in Postmenopausal Osteoporosis.Appl Biochem Biotechnol. 2025 Jun;197(6):3982-4001. doi: 10.1007/s12010-025-05216-1. Epub 2025 Mar 25. Appl Biochem Biotechnol. 2025. PMID: 40131629
-
Identification of shared gene signatures and pathways for diagnosing osteoporosis with sarcopenia through integrated bioinformatics analysis and machine learning.BMC Musculoskelet Disord. 2024 Jun 3;25(1):435. doi: 10.1186/s12891-024-07555-2. BMC Musculoskelet Disord. 2024. PMID: 38831425 Free PMC article.
References
-
- Aoki Y., Furukawa K., Suzuki D., Takayashiki T., Kuboki S., Takano S., et al. (2022). Influence of sarcopenia as defined by EWGSOP2 on complications after pancreaticoduodenectomy and on the prognosis of pancreatic head cancer: A prospective cohort study. Nutrition 99-100, 111660. 10.1016/j.nut.2022.111660 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

