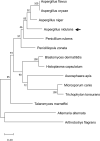
Homeobox transcription factor HbxA influences expression of over one thousand genes in the model fungus Aspergillus nidulans
- PMID: 37478074
- PMCID: PMC10361519
- DOI: 10.1371/journal.pone.0286271
Homeobox transcription factor HbxA influences expression of over one thousand genes in the model fungus Aspergillus nidulans
Abstract
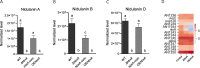
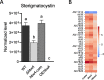
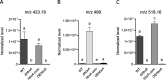
In fungi, conserved homeobox-domain proteins are transcriptional regulators governing development. In Aspergillus species, several homeobox-domain transcription factor genes have been identified, among them, hbxA/hbx1. For instance, in the opportunistic human pathogen Aspergillus fumigatus, hbxA is involved in conidial production and germination, as well as virulence and secondary metabolism, including production of fumigaclavines, fumiquinazolines, and chaetominine. In the agriculturally important fungus Aspergillus flavus, disruption of hbx1 results in fluffy aconidial colonies unable to produce sclerotia. hbx1 also regulates production of aflatoxins, cyclopiazonic acid and aflatrem. Furthermore, transcriptome studies revealed that hbx1 has a broad effect on the A. flavus genome, including numerous genes involved in secondary metabolism. These studies underline the importance of the HbxA/Hbx1 regulator, not only in developmental processes but also in the biosynthesis of a broad number of fungal natural products, including potential medical drugs and mycotoxins. To gain further insight into the regulatory scope of HbxA in Aspergilli, we studied its role in the model fungus Aspergillus nidulans. Our present study of the A. nidulans hbxA-dependent transcriptome revealed that more than one thousand genes are differentially expressed when this regulator was not transcribed at wild-type levels, among them numerous transcription factors, including those involved in development as well as in secondary metabolism regulation. Furthermore, our metabolomics analyses revealed that production of several secondary metabolites, some of them associated with A. nidulans hbxA-dependent gene clusters, was also altered in deletion and overexpression hbxA strains compared to the wild type, including synthesis of nidulanins A, B and D, versicolorin A, sterigmatocystin, austinol, dehydroaustinol, and three unknown novel compounds.
Copyright: © 2023 Pandit et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Oiartzabal-Arano E, Perez-de-Nanclares-Arregi E, Espeso EA, Etxebeste O. Apical control of conidiation in Aspergillus nidulans. Curr Genet. 2016. May 1;62(2):371–7. - PubMed
-
- Etxebeste O, Garzia A, Espeso EA, Ugalde U. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 2010. Dec;18(12):569–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

