Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants
- PMID: 37478179
- PMCID: PMC11803526
- DOI: 10.1126/sciadv.adg5953
Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants
Abstract
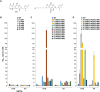
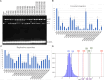
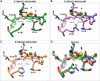
HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), first-line antiretroviral therapeutics widely used in the clinic. Resistance to even the best INSTIs is a problem, and the mechanisms of resistance are poorly understood. Here, we analyze combinations of the mutations E138K, G140A/S, and Q148H/K/R, which confer resistance to INSTIs. The investigational drug 4d more effectively inhibited the mutants compared with the approved drug Dolutegravir (DTG). We present 11 new cryo-EM structures of drug-resistant HIV-1 intasomes bound to DTG or 4d, with better than 3-Å resolution. These structures, complemented with free energy simulations, virology, and enzymology, explain the mechanisms of DTG resistance involving E138K + G140A/S + Q148H/K/R and show why 4d maintains potency better than DTG. These data establish a foundation for further development of INSTIs that potently inhibit resistant forms in integrase.
Figures


References
-
- D. O. Passos, M. Li, R. Craigie, D. Lyumkis, "Retroviral integrase: Structure, mechanism, and inhibition" in (Academic Press, 2021; https://sciencedirect.com/science/article/pii/S187460472100007X), vol. 50 of The Enzymes, pp. 249–300. - PMC - PubMed
-
- Engelman A., Mizuuchi K., Craigie R., HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell 67, 1211–1221 (1991). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

