Acetylation of the NS3 helicase by KAT5γ is essential for flavivirus replication
- PMID: 37478852
- PMCID: PMC10782998
- DOI: 10.1016/j.chom.2023.06.013
Acetylation of the NS3 helicase by KAT5γ is essential for flavivirus replication
Abstract
Direct targeting of essential viral enzymes such as proteases, polymerases, and helicases has long been the major focus of antiviral drug design. Although successful for some viral enzymes, targeting viral helicases is notoriously difficult to achieve, demanding alternative strategies. Here, we show that the NS3 helicase of Zika virus (ZIKV) undergoes acetylation in its RNA-binding tunnel. Regulation of the acetylated state of K389 in ZIKV NS3 modulates RNA binding and unwinding and is required for efficient viral replication. NS3 acetylation is mediated by a specific isoform of the host acetyltransferase KAT5 (KAT5γ), which translocates from the nucleus to viral replication complexes upon infection. NS3 acetylation by KAT5γ and its proviral role are also conserved in West Nile virus (WNV), dengue virus (DENV), and yellow fever virus (YFV). Our study provides molecular insight into how a cellular acetyltransferase regulates viral helicase functions, unveiling a previously unknown target for antiviral drug development.
Keywords: NS3 protein; Zika virus; acetylation; flaviviruses; post-translational modifications; viral helicase.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

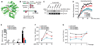

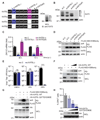
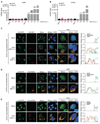
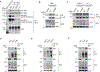
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

