Monitoring Both Extended and Tryptic Forms of Stable Isotope-Labeled Standard Peptides Provides an Internal Quality Control of Proteolytic Digestion in Targeted Mass Spectrometry-Based Assays
- PMID: 37478973
- PMCID: PMC10458721
- DOI: 10.1016/j.mcpro.2023.100621
Monitoring Both Extended and Tryptic Forms of Stable Isotope-Labeled Standard Peptides Provides an Internal Quality Control of Proteolytic Digestion in Targeted Mass Spectrometry-Based Assays
Abstract
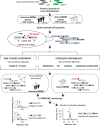
Targeted mass spectrometry (MS)-based proteomic assays, such as multiplexed multiple reaction monitoring (MRM)-MS assays, enable sensitive and specific quantification of proteotypic peptides as stoichiometric surrogates for proteins. Efforts are underway to expand the use of MRM-MS assays in clinical environments, which requires a reliable strategy to monitor proteolytic digestion efficiency within individual samples. Towards this goal, extended stable isotope-labeled standard (SIS) peptides (hE), which incorporate native proteolytic cleavage sites, can be spiked into protein lysates prior to proteolytic (trypsin) digestion, and release of the tryptic SIS peptide (hT) can be monitored. However, hT measurements alone cannot monitor the extent of digestion and may be confounded by matrix effects specific to individual patient samples; therefore, they are not sufficient to monitor sample-to-sample digestion variability. We hypothesized that measuring undigested hE, along with its paired hT, would improve detection of digestion issues compared to only measuring hT. We tested the ratio of the SIS pair measurements, or hE/hT, as a quality control (QC) metric of trypsin digestion for two MRM assays: a direct-MRM (398 targets) and an immuno-MRM (126 targets requiring immunoaffinity peptide enrichment) assay, with extended SIS peptides observable for 54% (216) and 62% (78) of the targets, respectively. We evaluated the quantitative bias for each target in a series of experiments that adversely affected proteolytic digestion (e.g., variable digestion times, pH, and temperature). We identified a subset of SIS pairs (36 for the direct-MRM, 7 for the immuno-MRM assay) for which the hE/hT ratio reliably detected inefficient digestion that resulted in decreased assay sensitivity and unreliable endogenous quantification. The hE/hT ratio was more responsive to a decrease in digestion efficiency than a metric based on hT measurements alone. For clinical-grade MRM-MS assays, this study describes a ready-to-use QC panel and also provides a road map for designing custom QC panels.
Keywords: clinical proteomics; immunoaffinity enrichment; quality control; quantification; targeted mass spectrometry; trypsin digestion.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures



References
-
- Picotti P., Bodenmiller B., Aebersold R. Proteomics meets the scientific method. Nat. Methods. 2013;10:24–27. - PubMed
-
- Neubert H., Shuford C.M., Olah T.V., Garofolo F., Schultz G.A., Jones B.R., et al. Protein biomarker quantification by immunoaffinity liquid chromatography-tandem mass spectrometry: current state and future vision. Clin. Chem. 2020;66:282–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

