The STROMICS genome study: deep whole-genome sequencing and analysis of 10K Chinese patients with ischemic stroke reveal complex genetic and phenotypic interplay
- PMID: 37479695
- PMCID: PMC10362040
- DOI: 10.1038/s41421-023-00582-8
The STROMICS genome study: deep whole-genome sequencing and analysis of 10K Chinese patients with ischemic stroke reveal complex genetic and phenotypic interplay
Abstract
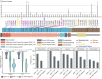
Ischemic stroke is a leading cause of global mortality and long-term disability. However, there is a paucity of whole-genome sequencing studies on ischemic stroke, resulting in limited knowledge of the interplay between genomic and phenotypic variations among affected patients. Here, we outline the STROMICS design and present the first whole-genome analysis on ischemic stroke by deeply sequencing and analyzing 10,241 stroke patients from China. We identified 135.59 million variants, > 42% of which were novel. Notable disparities in allele frequency were observed between Chinese and other populations for 89 variants associated with stroke risk and 10 variants linked to response to stroke medications. We investigated the population structure of the participants, generating a map of genetic selection consisting of 31 adaptive signals. The adaption of the MTHFR rs1801133-G allele, which links to genetically evaluated VB9 (folate acid) in southern Chinese patients, suggests a gene-specific folate supplement strategy. Through genome-wide association analysis of 18 stroke-related traits, we discovered 10 novel genetic-phenotypic associations and extensive cross-trait pleiotropy at 6 lipid-trait loci of therapeutic relevance. Additionally, we found that the set of loss-of-function and cysteine-altering variants present in the causal gene NOTCH3 for the autosomal dominant stroke disorder CADASIL displayed a broad neuro-imaging spectrum. These findings deepen our understanding of the relationship between the population and individual genetic layout and clinical phenotype among stroke patients, and provide a foundation for future efforts to utilize human genetic knowledge to investigate mechanisms underlying ischemic stroke outcomes, discover novel therapeutic targets, and advance precision medicine.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

