Predicting disease severity in metachromatic leukodystrophy using protein activity and a patient phenotype matrix
- PMID: 37480112
- PMCID: PMC10360315
- DOI: 10.1186/s13059-023-03001-z
Predicting disease severity in metachromatic leukodystrophy using protein activity and a patient phenotype matrix
Abstract
Background: Metachromatic leukodystrophy (MLD) is a lysosomal storage disorder caused by mutations in the arylsulfatase A gene (ARSA) and categorized into three subtypes according to age of onset. The functional effect of most ARSA mutants remains unknown; better understanding of the genotype-phenotype relationship is required to support newborn screening (NBS) and guide treatment.
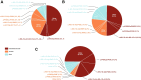
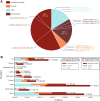
Results: We collected a patient data set from the literature that relates disease severity to ARSA genotype in 489 individuals with MLD. Patient-based data were used to develop a phenotype matrix that predicts MLD phenotype given ARSA alleles in a patient's genotype with 76% accuracy. We then employed a high-throughput enzyme activity assay using mass spectrometry to explore the function of ARSA variants from the curated patient data set and the Genome Aggregation Database (gnomAD). We observed evidence that 36% of variants of unknown significance (VUS) in ARSA may be pathogenic. By classifying functional effects for 251 VUS from gnomAD, we reduced the incidence of genotypes of unknown significance (GUS) by over 98.5% in the overall population.
Conclusions: These results provide an additional tool for clinicians to anticipate the disease course in MLD patients, identifying individuals at high risk of severe disease to support treatment access. Our results suggest that more than 1 in 3 VUS in ARSA may be pathogenic. We show that combining genetic and biochemical information increases diagnostic yield. Our strategy may apply to other recessive diseases, providing a tool to address the challenge of interpreting VUS within genotype-phenotype relationships and NBS.
Keywords: Genotype–phenotype relationship; Mass spectrometry; Metachromatic leukodystrophy; Mutation; Newborn screening; Phenotype.
© 2023. The Author(s).
Conflict of interest statement
MT, JS, and HPN were fulltime employees of BioMarin Pharmaceutical Inc, during the conduct of the study. SF, JHL, and WTC are fulltime employees of BioMarin Pharmaceutical Inc. XH, HPN, MC, DS, TS, and MHG declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

