Altered Expression of Heme Oxygenase 2 in Heme Oxygenase 1-deficient Mouse Embryos
- PMID: 37480265
- PMCID: PMC10424577
- DOI: 10.1369/00221554231189310
Altered Expression of Heme Oxygenase 2 in Heme Oxygenase 1-deficient Mouse Embryos
Abstract
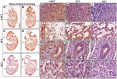
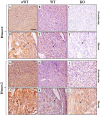
Heme oxygenases (Hmoxs) are enzymes that catalyze the first and rate-limiting step in the degradation of heme to carbon monoxide, iron, and biliverdin. The two main isozymes, namely Hmox1 and Hmox2, are encoded by two different genes. Mutation of the Hmox1 gene in mice is known to cause extensive prenatal lethality, and limited information is available about the expression of Hmox proteins in developing mouse embryos. In this study, immunohistochemistry was used to perform a detailed investigation comparing Hmox proteins in Hmox1 wild-type and knockout (KO) mouse embryos collected from wild-type and heterozygous timed-matings. Western analysis for Hmoxs was also done in the organs of late-gestation embryos. The results demonstrated cytoplasmic and nuclear localization of Hmoxs in all the organs examined in wild-type embryos. Interestingly, Hmox2 immunoreactive protein signals were significantly low in most of the organs of mid- and late-gestation Hmox1-KO embryos. Furthermore, relative levels of Hmox2 were revealed to be significantly lower in the lung and kidney of late-gestation Hmox1-KO embryos by western analysis, which complemented the immunohistochemistry findings in these two organs. The current study provides detailed immunoexpression patterns of Hmox proteins in wild-type and Hmox1-KO mouse embryos in mid- and late-gestation.
Keywords: heme oxygenase; immunohistochemistry; knockout; mouse embryos; phenotype.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures









Similar articles
-
Expression of angiogenic factors in the placenta of heme oxygenase-1 deficient mouse embryo.Reprod Biol. 2023 Dec;23(4):100822. doi: 10.1016/j.repbio.2023.100822. Epub 2023 Nov 17. Reprod Biol. 2023. PMID: 37979494
-
Heme Oxygenase 1 and 2 Differentially Regulate Glucose Metabolism and Adipose Tissue Mitochondrial Respiration: Implications for Metabolic Dysregulation.Int J Mol Sci. 2020 Sep 27;21(19):7123. doi: 10.3390/ijms21197123. Int J Mol Sci. 2020. PMID: 32992485 Free PMC article.
-
Heme Oxygenases in Cardiovascular Health and Disease.Physiol Rev. 2016 Oct;96(4):1449-508. doi: 10.1152/physrev.00003.2016. Physiol Rev. 2016. PMID: 27604527 Free PMC article. Review.
-
An association between polymorphism of the heme oxygenase-1 and -2 genes and age-related macular degeneration.Mol Biol Rep. 2012 Mar;39(3):2081-7. doi: 10.1007/s11033-011-0955-3. Epub 2011 Jun 7. Mol Biol Rep. 2012. PMID: 21647550 Free PMC article.
-
The clinical relevance of heme detoxification by the macrophage heme oxygenase system.Front Immunol. 2024 Mar 22;15:1379967. doi: 10.3389/fimmu.2024.1379967. eCollection 2024. Front Immunol. 2024. PMID: 38585264 Free PMC article. Review.
Cited by
-
Heme oxygenase/carbon monoxide system and development of the heart.Med Gas Res. 2025 Mar 1;15(1):10-22. doi: 10.4103/mgr.MEDGASRES-D-24-00031. Epub 2024 Sep 25. Med Gas Res. 2025. PMID: 39324891 Free PMC article. Review.
-
The Role of Ferroptosis in Amyotrophic Lateral Sclerosis Treatment.Neurochem Res. 2024 Oct;49(10):2653-2667. doi: 10.1007/s11064-024-04194-w. Epub 2024 Jun 12. Neurochem Res. 2024. PMID: 38864944 Review.
References
-
- Cruse I, Maines MD. Evidence suggesting that the two forms of heme oxygenase are products of different genes. J Biol Chem. 1988;263(7):3348–53. - PubMed
-
- Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86(2):583–650. - PubMed
-
- McCoubrey WK, Jr, Huang TJ, Maines MD. Isolation and characterization of a cDNA from the rat brain that encodes hemoprotein heme oxygenase-3. Eur J Biochem. 1997;247(2):725–32. - PubMed
-
- Hayashi S, Omata Y, Sakamoto H, Higashimoto Y, Hara T, Sagara Y, Noguchi M. Characterization of rat heme oxygenase-3 gene. Implication of processed pseudogenes derived from heme oxygenase-2 gene. Gene. 2004;336(2):241–50. - PubMed
-
- Maines MD, Trakshel GM, Kutty RK. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. J Biol Chem. 1986;261(1):411–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

