A comprehensive Drosophila resource to identify key functional interactions between SARS-CoV-2 factors and host proteins
- PMID: 37480566
- PMCID: PMC10962759
- DOI: 10.1016/j.celrep.2023.112842
A comprehensive Drosophila resource to identify key functional interactions between SARS-CoV-2 factors and host proteins
Abstract
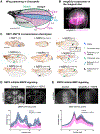
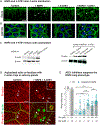
Development of effective therapies against SARS-CoV-2 infections relies on mechanistic knowledge of virus-host interface. Abundant physical interactions between viral and host proteins have been identified, but few have been functionally characterized. Harnessing the power of fly genetics, we develop a comprehensive Drosophila COVID-19 resource (DCR) consisting of publicly available strains for conditional tissue-specific expression of all SARS-CoV-2 encoded proteins, UAS-human cDNA transgenic lines encoding established host-viral interacting factors, and GAL4 insertion lines disrupting fly homologs of SARS-CoV-2 human interacting proteins. We demonstrate the utility of the DCR to functionally assess SARS-CoV-2 genes and candidate human binding partners. We show that NSP8 engages in strong genetic interactions with several human candidates, most prominently with the ATE1 arginyltransferase to induce actin arginylation and cytoskeletal disorganization, and that two ATE1 inhibitors can reverse NSP8 phenotypes. The DCR enables parallel global-scale functional analysis of SARS-CoV-2 components in a prime genetic model system.
Keywords: ATE1; CP: Microbiology; DRC; Drosophila; Drosophila Covid-19 resource; NSP8; NSPs; Orf3a; SARS-CoV-2; arginylation; human interactors; non-structural proteins.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.B. has equity interests in Synbal Inc., a company that may potentially benefit from the research results. E.B. also serves on the Board of Directors and Scientific Advisory Board of Synbal. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies.
Figures





References
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, et al. (2020). Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19. 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

