Dual-locus, dual-HDR editing permits efficient generation of antigen-specific regulatory T cells with robust suppressive activity
- PMID: 37481700
- PMCID: PMC10556186
- DOI: 10.1016/j.ymthe.2023.07.016
Dual-locus, dual-HDR editing permits efficient generation of antigen-specific regulatory T cells with robust suppressive activity
Abstract
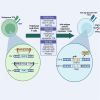
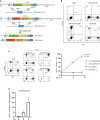
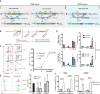
Adoptive regulatory T (Treg) cell therapy is predicted to modulate immune tolerance in autoimmune diseases, including type 1 diabetes (T1D). However, the requirement for antigen (ag) specificity to optimally orchestrate tissue-specific, Treg cell-mediated tolerance limits effective clinical application. To address this challenge, we present a single-step, combinatorial gene editing strategy utilizing dual-locus, dual-homology-directed repair (HDR) to generate and specifically expand ag-specific engineered Treg (EngTreg) cells derived from donor CD4+ T cells. Concurrent delivery of CRISPR nucleases and recombinant (r)AAV homology donor templates targeting FOXP3 and TRAC was used to achieve three parallel goals: enforced, stable expression of FOXP3; replacement of the endogenous T cell receptor (TCR) with an islet-specific TCR; and selective enrichment of dual-edited cells. Each HDR donor template contained an alternative component of a heterodimeric chemically inducible signaling complex (CISC), designed to activate interleukin-2 (IL-2) signaling in response to rapamycin, promoting expansion of only dual-edited EngTreg cells. Using this approach, we generated purified, islet-specific EngTreg cells that mediated robust direct and bystander suppression of effector T (Teff) cells recognizing the same or a different islet antigen peptide, respectively. This platform is broadly adaptable for use with alternative TCRs or other targeting moieties for application in tissue-specific autoimmune or inflammatory diseases.
Keywords: AAV6; CISC; CRISPR; Treg; ag-specific; dual-HDR; enrichment.
Copyright © 2023 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.J.R. is a scientific co-founder and scientific advisory board (SAB) member of GentiBio, Inc. and scientific co-Founder and SAB member of BeBiopharma, Inc. D.J.R. received past and current funding from GentiBio, Inc. for related work. J.B. is a scientific co-founder and SAB member of GentiBio, Inc. J.B. received past and current funding from GentiBio, Inc. for related work. D.J.R., P.J.C., and J.B. are inventors on patents describing methods for generating ag-specific engineered regulatory T cells and/or use of the CISC platform.
Figures



References
-
- Honaker Y., Hubbard N., Xiang Y., Fisher L., Hagin D., Sommer K., Song Y., Yang S.J., Lopez C., Tappen T., et al. Gene editing to induce FOXP3 expression in human CD4(+) T cells leads to a stable regulatory phenotype and function. Sci. Transl. Med. 2020;12 - PubMed
-
- Yang S.J., Singh A.K., Drow T., Tappen T., Honaker Y., Barahmand-Pour-Whitman F., Linsley P.S., Cerosaletti K., Mauk K., Xiang Y., et al. Pancreatic islet-specific engineered T(regs) exhibit robust antigen-specific and bystander immune suppression in type 1 diabetes models. Sci. Transl. Med. 2022;14 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

