Hydrogel-encapsulation to enhance bacterial diagnosis of colon inflammation
- PMID: 37481834
- PMCID: PMC10792543
- DOI: 10.1016/j.biomaterials.2023.122246
Hydrogel-encapsulation to enhance bacterial diagnosis of colon inflammation
Abstract
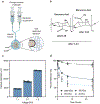
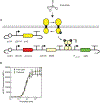
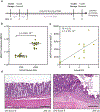
Bacteria can be genetically programmed to sense and report the presence of disease biomarkers in the gastrointestinal (GI) tract. However, diagnostic bacteria are typically delivered via oral administration of liquid cultures, resulting in poor survival and high dispersal in vivo. These limitations confound recovery and analysis of engineered bacteria from GI or stool samples. Here, we demonstrate that encapsulating bacteria inside of alginate core-shell particles enables robust survival, containment, and diagnostic function in vivo. We demonstrate these benefits by encapsulating a strain engineered to report the presence of the biomarker thiosulfate via fluorescent protein expression in order to diagnose dextran sodium sulfate-induced colitis in rats. Hydrogel-encapsulated bacteria engineered to sense and respond to physiological stimuli should enable minimally invasive monitoring of a wide range of diseases and have applications as next-generation smart therapeutics.
Keywords: Colitis; Diagnostic bacteria; Hydrogel encapsulation; Synthetic biology.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Omid Veiseh and Jeffrey Tabor reports a relationship with Pana Bio that includes: board membership and equity or stocks. Samira Aghlara-Fotovat, Elena Musteata, Michael D. Doerfert, Moshe Baruch, Jeffrey J. Tabor, Omid Veiseh has patent pending to Pana Bio. Moshe Baruch is current employee of Pana Bio.
Figures
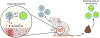


References
-
- Sender R, Fuchs S & Milo R Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164, 337–340 (2016). - PubMed
-
- Ley RE, Peterson DA & Gordon JI Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006). - PubMed
-
- Cryan JF, O’Riordan KJ, Sandhu K, Peterson V & Dinan TG The gut microbiome in neurological disorders. Lancet Neurol. 19, 179–194 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

