Molecular profiling of solid tumors by next-generation sequencing: an experience from a clinical laboratory
- PMID: 37483495
- PMCID: PMC10359709
- DOI: 10.3389/fonc.2023.1208244
Molecular profiling of solid tumors by next-generation sequencing: an experience from a clinical laboratory
Abstract
Background: Personalized targeted therapies have transformed management of several solid tumors. Timely and accurate detection of clinically relevant genetic variants in tumor is central to the implementation of molecular targeted therapies. To facilitate precise molecular testing in solid tumors, targeted next-generation sequencing (NGS) assays have emerged as a valuable tool. In this study, we provide an overview of the technical validation, diagnostic yields, and spectrum of variants observed in 3,164 solid tumor samples that were tested as part of the standard clinical diagnostic assessment in an academic healthcare institution over a period of 2 years.
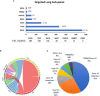
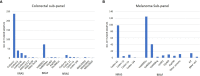
Methods: The Ion Ampliseq™ Cancer Hotspot Panel v2 assay (ThermoFisher) that targets ~2,800 COSMIC mutations from 50 oncogenes and tumor suppressor genes was validated, and a total of 3,164 tumor DNA samples were tested in 2 years. A total of 500 tumor samples were tested by the comprehensive panel containing all the 50 genes. Other samples, including 1,375 lung cancer, 692 colon cancer, 462 melanoma, and 135 brain cancer, were tested by tumor-specific targeted subpanels including a few clinically actionable genes.
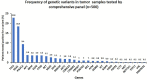
Results: Of 3,164 patient samples, 2,016 (63.7%) tested positive for at least one clinically relevant variant. Of 500 samples tested by a comprehensive panel, 290 had a clinically relevant variant with TP53, KRAS, and PIK3CA being the most frequently mutated genes. The diagnostic yields in major tumor types were as follows: breast (58.4%), colorectal (77.6%), lung (60.4%), pancreatic (84.6%), endometrial (72.4%), ovary (57.1%), and thyroid (73.9%). Tumor-specific targeted subpanels also demonstrated high diagnostic yields: lung (69%), colon (61.2%), melanoma (69.7%), and brain (20.7%). Co-occurrence of mutations in more than one gene was frequently observed.
Conclusions: The findings of our study demonstrate the feasibility of integrating an NGS-based gene panel screen as part of a standard diagnostic protocol for solid tumor assessment. High diagnostic rates enable significant clinical impact including improved diagnosis, prognosis, and clinical management in patients with solid tumors.
Keywords: NGS; colon cancer; lung cancer; melanoma; solid tumor.
Copyright © 2023 Bhai, Turowec, Santos, Kerkhof, Pickard, Foroutan, Breadner, Cecchini, Levy, Stuart, Welch, Howlett, Lin and Sadikovic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

