HURP localization in metaphase is the result of a multi-step process requiring its phosphorylation at Ser627 residue
- PMID: 37484914
- PMCID: PMC10361663
- DOI: 10.3389/fcell.2023.981425
HURP localization in metaphase is the result of a multi-step process requiring its phosphorylation at Ser627 residue
Abstract
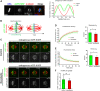
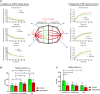
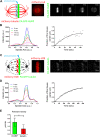
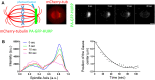
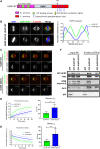
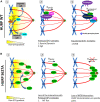
Faithful chromosome segregation during cell division requires accurate mitotic spindle formation. As mitosis occurs rapidly within the cell cycle, the proteins involved in mitotic spindle assembly undergo rapid changes, including their interactions with other proteins. The proper localization of the HURP protein on the kinetochore fibers, in close proximity to chromosomes, is crucial for ensuring accurate congression and segregation of chromosomes. In this study, we employ photoactivation and FRAP experiments to investigate the impact of alterations in microtubule flux and phosphorylation of HURP at the Ser627 residue on its dynamics. Furthermore, through immunoprecipitations assays, we demonstrate the interactions of HURP with various proteins, such as TPX2, Aurora A, Eg5, Dynein, Kif5B, and Importin β, in mammalian cells during mitosis. We also find that phosphorylation of HURP at Ser627 regulates its interaction with these partners during mitosis. Our findings suggest that HURP participates in at least two distinct complexes during metaphase to ensure its proper localization in close proximity to chromosomes, thereby promoting the bundling and stabilization of kinetochore fibers.
Keywords: Aurora A; FRAP (fluorescence recovery after photobleaching); HURP dynamics; HURP localization; HURP phosphorylation; TPX2; mitosis; photoactivation.
Copyright © 2023 Didaskalou, Efstathiou, Galtsidis, Kesisova, Halavatyi, Elmali, Tsolou, Girod and Koffa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

