Non-specific recognition of histone modifications by H3K9bhb antibody
- PMID: 37485368
- PMCID: PMC10362303
- DOI: 10.1016/j.isci.2023.107235
Non-specific recognition of histone modifications by H3K9bhb antibody
Abstract
Ketone bodies are short-chain fatty acids produced in the liver during periods of limited glucose availability that provide an alternative energy source for the brain, heart, and skeletal muscle. Beyond this metabolic role, β-hydroxybutyrate (BHB), is gaining recognition as a signaling molecule. Lysine β-hydroxybutyrylation (Kbhb) is a newly discovered post-translational modification in which BHB is covalently attached to lysine ε-amino groups. This protein adduct is metabolically sensitive, dependent on BHB concentration, and found on proteins in multiple intracellular compartments. Therefore, Kbhb is hypothesized to be an important component of ketone body-regulated physiology. Kbhb on histones is proposed to be an epigenetic regulator, which links metabolic alterations to gene expression. However, we found that the widely used antibody against β-hydroxybutyrylated lysine 9 on histone H3 (H3K9bhb) also recognizes other modification(s) that likely include acetylation. Therefore, caution must be used when interpreting gene regulation data acquired with the H3K9bhb antibody.
Keywords: Biochemistry; Biological sciences; Cell biology; Molecular biology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
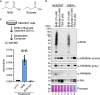

Update of
-
Non-specific recognition of histone modifications by H3K9bhb antibody.bioRxiv [Preprint]. 2023 Apr 13:2023.04.12.536655. doi: 10.1101/2023.04.12.536655. bioRxiv. 2023. Update in: iScience. 2023 Jun 29;26(7):107235. doi: 10.1016/j.isci.2023.107235. PMID: 37090555 Free PMC article. Updated. Preprint.

