Non-homologous end-joining-deficient filamentous fungal strains mitigate the impact of off-target mutations during the application of CRISPR/Cas9
- PMID: 37486124
- PMCID: PMC10470509
- DOI: 10.1128/mbio.00668-23
Non-homologous end-joining-deficient filamentous fungal strains mitigate the impact of off-target mutations during the application of CRISPR/Cas9
Abstract
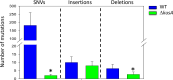
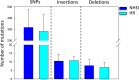
CRISPR/Cas9 genome editing technology has been implemented in almost all living organisms. Its editing precision appears to be very high and therefore could represent a big change from conventional genetic engineering approaches. However, guide RNA binding to nucleotides similar to the target site could result in undesired off-target mutations. Despite this, evaluating whether mutations occur is rarely performed in genome editing studies. In this study, we generated CRISPR/Cas9-derived filamentous fungal strains and analyzed them for the occurrence of mutations, and to which extent genome stability affects their occurrence. As a test case, we deleted the (hemi-)cellulolytic regulator-encoding gene xlnR in two Aspergillus niger strains: a wild type (WT) and a non-homologous end-joining (NHEJ)-deficient strain ΔkusA. Initial phenotypic analysis suggested a much higher prevalence of mutations in the WT compared to NHEJ-deficient strains, which was confirmed and quantified by whole-genome sequencing analysis. Our results clearly demonstrate that CRISPR/Cas9 applied to an NHEJ-deficient strain is an efficient strategy to avoid unwanted mutations. IMPORTANCE Filamentous fungi are commonly used biofactories for the production of industrially relevant proteins and metabolites. Often, fungal biofactories undergo genetic development (genetic engineering, genome editing, etc.) aimed at improving production yields. In this context, CRISPR/Cas9 has gained much attention as a genome editing strategy due to its simplicity, versatility, and precision. However, despite the high level of accuracy reported for CRISPR/Cas9, in some cases unintentional cleavages in non-targeted loci-known as off-target mutations-could arise. While biosafety should be a central feature of emerging biotechnologies to minimize unintended consequences, few studies quantitatively evaluate the risk of off-target mutations. This study demonstrates that the use of non-homologous end-joining-deficient fungal strains drastically reduces the number of unintended genomic mutations, ensuring that CRISPR/Cas9 can be safely applied for strain development.
Keywords: Aspergillus niger; CRISPR-Cas9; biosafety; risk assessment; ΔkusA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass.Enzyme Microb Technol. 2020 May;136:109508. doi: 10.1016/j.enzmictec.2020.109508. Epub 2020 Jan 24. Enzyme Microb Technol. 2020. PMID: 32331715
-
Rapid and marker-free gene replacement in citric acid-producing Aspergillus tubingensis (A. niger) WU-2223L by the CRISPR/Cas9 system-based genome editing technique using DNA fragments encoding sgRNAs.J Biosci Bioeng. 2021 Jun;131(6):579-588. doi: 10.1016/j.jbiosc.2021.01.011. Epub 2021 Feb 19. J Biosci Bioeng. 2021. PMID: 33612423
-
[An efficient marker-free genome editing method for Aspergillus niger].Sheng Wu Gong Cheng Xue Bao. 2022 Dec 25;38(12):4744-4755. doi: 10.13345/j.cjb.220162. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 36593207 Chinese.
-
Application of CRISPR in Filamentous Fungi and Macrofungi: From Component Function to Development Potentiality.ACS Synth Biol. 2023 Jul 21;12(7):1908-1923. doi: 10.1021/acssynbio.3c00099. Epub 2023 Jul 5. ACS Synth Biol. 2023. PMID: 37404005 Review.
-
The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans.J Genet Genomics. 2015 Aug 20;42(8):413-21. doi: 10.1016/j.jgg.2015.06.005. Epub 2015 Jun 26. J Genet Genomics. 2015. PMID: 26336798 Free PMC article. Review.
Cited by
-
Regulation and induction of fungal secondary metabolites: a comprehensive review.Arch Microbiol. 2025 Jul 1;207(8):189. doi: 10.1007/s00203-025-04386-0. Arch Microbiol. 2025. PMID: 40590991 Review.
-
Competition between homologous chromosomal DNA and exogenous donor DNA to repair CRISPR/Cas9-induced double-strand breaks in Aspergillus niger.Fungal Biol Biotechnol. 2024 Oct 15;11(1):15. doi: 10.1186/s40694-024-00184-3. Fungal Biol Biotechnol. 2024. PMID: 39407321 Free PMC article.
-
Oligonucleotide-based CRISPR-Cas9 toolbox for efficient engineering of Komagataella phaffii.FEMS Yeast Res. 2024 Jan 9;24:foae026. doi: 10.1093/femsyr/foae026. FEMS Yeast Res. 2024. PMID: 39179418 Free PMC article.
-
Increasing the efficiency of CRISPR/Cas9-mediated genome editing in the citrus postharvest pathogen Penicillium digitatum.Fungal Biol Biotechnol. 2024 Jul 13;11(1):8. doi: 10.1186/s40694-024-00179-0. Fungal Biol Biotechnol. 2024. PMID: 39003486 Free PMC article.
-
NHEJ and HDR can occur simultaneously during gene integration into the genome of Aspergillus niger.Fungal Biol Biotechnol. 2024 Aug 5;11(1):10. doi: 10.1186/s40694-024-00180-7. Fungal Biol Biotechnol. 2024. PMID: 39103967 Free PMC article.
References
-
- Meyer V. 2021. Metabolic engineering of Filamentous fungi, p 765–801. In Nielsen J, Stephanopoulos G, Lee SY (ed), Metabolic engineering. doi:10.1002/9783527823468 - DOI
-
- Álvarez-Escribano I, Sasse C, Bok JW, Na H, Amirebrahimi M, Lipzen A, Schackwitz W, Martin J, Barry K, Gutiérrez G, Cea-Sánchez S, Marcos AT, Grigoriev IV, Keller NP, Braus GH, Cánovas D. 2019. Genome sequencing of evolved Aspergilli populations reveals robust genomes, transversions in A. flavus, and sexual aberrancy in non-homologous end-joining Mutants. BMC Biol 17:88. doi:10.1186/s12915-019-0702-0 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

