A combined microscopy and single-cell sequencing approach reveals the ecology, morphology, and phylogeny of uncultured lineages of zoosporic fungi
- PMID: 37486265
- PMCID: PMC10470594
- DOI: 10.1128/mbio.01313-23
A combined microscopy and single-cell sequencing approach reveals the ecology, morphology, and phylogeny of uncultured lineages of zoosporic fungi
Abstract
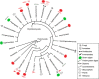
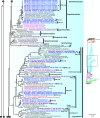
Environmental DNA analyses of fungal communities typically reveal a much larger diversity than can be ascribed to known species. Much of this hidden diversity lies within undescribed fungal lineages, especially the early diverging fungi (EDF). Although these EDF often represent new lineages even at the phylum level, they have never been cultured, making their morphology and ecology uncertain. One of the methods to characterize these uncultured fungi is a single-cell DNA sequencing approach. In this study, we established a large data set of single-cell sequences of EDF by manually isolating and photographing parasitic fungi on various hosts such as algae, protists, and micro-invertebrates, combined with subsequent long-read sequencing of the ribosomal DNA locus (rDNA). We successfully obtained rDNA sequences of 127 parasitic fungal cells, which clustered into 71 phylogenetic lineages belonging to seven phylum-level clades of EDF: Blastocladiomycota, Chytridiomycota, Aphelidiomycota, Rozellomycota, and three unknown phylum-level clades. Most of our single cells yielded novel sequences distinguished from both described taxa and existing metabarcoding data, indicating an expansive and hidden diversity of parasitic taxa of EDF. We also revealed an unexpected diversity of endobiotic Olpidium-like chytrids and hyper-parasitic lineages. Overall, by combining photographs of parasitic fungi with phylogenetic analyses, we were able to better understand the ecological function and morphology of many of the branches on the fungal tree of life known only from DNA sequences. IMPORTANCE Much of the diversity of microbes from natural habitats, such as soil and freshwater, comprise species and lineages that have never been isolated into pure culture. In part, this stems from a bias of culturing in favor of saprotrophic microbes over the myriad symbiotic ones that include parasitic and mutualistic relationships with other taxa. In the present study, we aimed to shed light on the ecological function and morphology of the many undescribed lineages of aquatic fungi by individually isolating and sequencing molecular barcodes from 127 cells of host-associated fungi using single-cell sequencing. By adding these sequences and their photographs into the fungal tree, we were able to understand the morphology of reproductive and vegetative structures of these novel fungi and to provide a hypothesized ecological function for them. These individual host-fungal cells revealed themselves to be complex environments despite their small size; numerous samples were hyper-parasitized with other zoosporic fungal lineages such as Rozellomycota.
Keywords: early diverging fungi; parasite; phylogeny; single-cell analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson K-H, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K. 2014. Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

