Genome-wide phylodynamic approach reveals the epidemic dynamics of the main Mycoplasma bovis subtype circulating in France
- PMID: 37486749
- PMCID: PMC10438803
- DOI: 10.1099/mgen.0.001067
Genome-wide phylodynamic approach reveals the epidemic dynamics of the main Mycoplasma bovis subtype circulating in France
Abstract
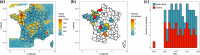
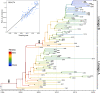
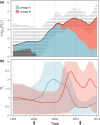
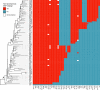
Mycoplasma bovis is a major aetiological agent of bovine respiratory disease worldwide. Genome-based analyses are increasingly being used to monitor the genetic diversity and global distribution of M. bovis, complementing existing subtyping schemes based on locus sequencing. However, these analyses have so far provided limited information on the spatiotemporal and population dynamics of circulating subtypes. Here we applied a genome-wide phylodynamic approach to explore the epidemic dynamics of 88 French M. bovis strains collected between 2000 and 2019 in France and belonging to the currently dominant polC subtype 2 (st2). A strong molecular clock signal detected in the genomic data enabled robust phylodynamic inferences, which estimated that the M. bovis st2 population in France is composed of two lineages that successively emerged from independent introductions of international strains. The first lineage appeared around 2000 and supplanted the previously established antimicrobial-susceptible polC subtype 1. The second lineage, which is likely more transmissible, progressively replaced the first M. bovis st2 lineage population from 2005 onward and became predominant after 2010. Analyses also showed a brief decline in this second M. bovis st2 lineage population in around 2011, possibly due to the challenge from the concurrent emergence of M. bovis polC subtype 3 in France. Finally, we identified non-synonymous mutations in genes associated with lineages, which raises prospects for identifying new surveillance molecular markers. A genome-wide phylodynamic approach provides valuable resources for monitoring the evolution and epidemic dynamics of circulating M. bovis subtypes, and may prove critical for developing more effective surveillance systems and disease control strategies.
Keywords: bacteria; cattle; fitness; lineage replacement; respiratory disease; surveillance.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

