Hepatocytes reprogram liver macrophages involving control of TGF-β activation, influencing liver regeneration and injury
- PMID: 37486964
- PMCID: PMC10368377
- DOI: 10.1097/HC9.0000000000000208
Hepatocytes reprogram liver macrophages involving control of TGF-β activation, influencing liver regeneration and injury
Abstract
Background: Macrophages play an important role in maintaining liver homeostasis and regeneration. However, it is not clear to what extent the different macrophage populations of the liver differ in terms of their activation state and which other liver cell populations may play a role in regulating the same.
Methods: Reverse transcription PCR, flow cytometry, transcriptome, proteome, secretome, single cell analysis, and immunohistochemical methods were used to study changes in gene expression as well as the activation state of macrophages in vitro and in vivo under homeostatic conditions and after partial hepatectomy.
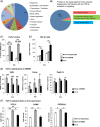
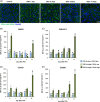
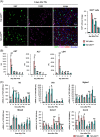
Results: We show that F4/80+/CD11bhi/CD14hi macrophages of the liver are recruited in a C-C motif chemokine receptor (CCR2)-dependent manner and exhibit an activation state that differs substantially from that of the other liver macrophage populations, which can be distinguished on the basis of CD11b and CD14 expressions. Thereby, primary hepatocytes are capable of creating an environment in vitro that elicits the same specific activation state in bone marrow-derived macrophages as observed in F4/80+/CD11bhi/CD14hi liver macrophages in vivo. Subsequent analyses, including studies in mice with a myeloid cell-specific deletion of the TGF-β type II receptor, suggest that the availability of activated TGF-β and its downregulation by a hepatocyte-conditioned milieu are critical. Reduction of TGF-βRII-mediated signal transduction in myeloid cells leads to upregulation of IL-6, IL-10, and SIGLEC1 expression, a hallmark of the activation state of F4/80+/CD11bhi/CD14hi macrophages, and enhances liver regeneration.
Conclusions: The availability of activated TGF-β determines the activation state of specific macrophage populations in the liver, and the observed rapid transient activation of TGF-β may represent an important regulatory mechanism in the early phase of liver regeneration in this context.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Julio Saez-Rodriguez has received funding from GSK, Pfizer and Sanofi, and fees/honoraria from Travere Therapeutics, Stadapharm, Astex, Pfizer and Grunenthal. The remaining authors have no conflicts to report.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

