Switching on cyclic di-GMP heterogeneity in Pseudomonas aeruginosa biofilms
- PMID: 37488256
- PMCID: PMC10752495
- DOI: 10.1038/s41564-023-01428-5
Switching on cyclic di-GMP heterogeneity in Pseudomonas aeruginosa biofilms
Abstract
Pseudomonas aeruginosa drives heterogeneity of cyclic di-GMP signalling in biofilms as a division-of-labour strategy to maximize colonization and dispersal using the protein HecE.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
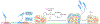
Comment on
-
A genetic switch controls Pseudomonas aeruginosa surface colonization.Nat Microbiol. 2023 Aug;8(8):1520-1533. doi: 10.1038/s41564-023-01403-0. Epub 2023 Jun 8. Nat Microbiol. 2023. PMID: 37291227
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

