Comparative pathogenicity of SARS-CoV-2 Omicron subvariants including BA.1, BA.2, and BA.5
- PMID: 37488344
- PMCID: PMC10366110
- DOI: 10.1038/s42003-023-05081-w
Comparative pathogenicity of SARS-CoV-2 Omicron subvariants including BA.1, BA.2, and BA.5
Abstract
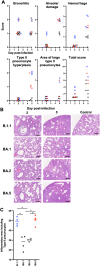
The unremitting emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants necessitates ongoing control measures. Given its rapid spread, the new Omicron subvariant BA.5 requires urgent characterization. Here, we comprehensively analyzed BA.5 with the other Omicron variants BA.1, BA.2, and ancestral B.1.1. Although in vitro growth kinetics of BA.5 was comparable among the Omicron subvariants, BA.5 was much more fusogenic than BA.1 and BA.2. Airway-on-a-chip analysis showed that, among Omicron subvariants, BA.5 had enhanced ability to disrupt the respiratory epithelial and endothelial barriers. Furthermore, in our hamster model, in vivo pathogenicity of BA.5 was slightly higher than that of the other Omicron variants and less than that of ancestral B.1.1. Notably, BA.5 gains efficient virus spread compared with BA.1 and BA.2, leading to prompt immune responses. Our findings suggest that BA.5 has low pathogenicity compared with the ancestral strain but enhanced virus spread /inflammation compared with earlier Omicron subvariants.
© 2023. The Author(s).
Conflict of interest statement
Y.Y. and T.N. are founders and shareholders of HiLung, Inc. J.K. is an employee of HiLung, Inc. Y.Y. is a co-inventor of a patent (PCT/JP2016/057254, “Method for inducing differentiation of alveolar epithelial cells”) related to this work. I.Y. reports speaker fees from Chugai Pharmaceutical Co, and AstraZeneca plt, outside the submitted work. The other authors declare no competing interests.
Figures




References
-
- WHO. "Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern (November 26, 2021)” https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.... (2020).
-
- Kislaya, I. C. et al. SARS-CoV-2 BA.5 vaccine breakthrough risk and severity compared with BA.2: a case-case and cohort study using Electronic Health Records in Portugal. medRxiv10.1101/2022.07.25.22277996 (2022).
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

