Zfp296 knockout enhances chromatin accessibility and induces a unique state of pluripotency in embryonic stem cells
- PMID: 37488353
- PMCID: PMC10366109
- DOI: 10.1038/s42003-023-05148-8
Zfp296 knockout enhances chromatin accessibility and induces a unique state of pluripotency in embryonic stem cells
Abstract
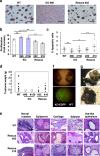
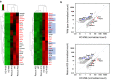
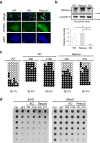
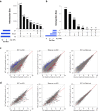
The Zfp296 gene encodes a zinc finger-type protein. Its expression is high in mouse embryonic stem cells (ESCs) but rapidly decreases following differentiation. Zfp296-knockout (KO) ESCs grew as flat colonies, which were reverted to rounded colonies by exogenous expression of Zfp296. KO ESCs could not form teratomas when transplanted into mice but could efficiently contribute to germline-competent chimeric mice following blastocyst injection. Transcriptome analysis revealed that Zfp296 deficiency up- and down-regulates a distinct group of genes, among which Dppa3, Otx2, and Pou3f1 were markedly downregulated. Chromatin immunoprecipitation sequencing demonstrated that ZFP296 binding is predominantly seen in the vicinity of the transcription start sites (TSSs) of a number of genes, and ZFP296 was suggested to negatively regulate transcription. Consistently, chromatin accessibility assay clearly showed that ZFP296 binding reduces the accessibility of the TSS regions of target genes. Zfp296-KO ESCs showed increased histone H3K9 di- and trimethylation. Co-immunoprecipitation analyses revealed interaction of ZFP296 with G9a and GLP. These results show that ZFP296 plays essential roles in maintaining the global epigenetic state of ESCs through multiple mechanisms including activation of Dppa3, attenuation of chromatin accessibility, and repression of H3K9 methylation, but that Zfp296-KO ESCs retain a unique state of pluripotency while lacking the teratoma-forming ability.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

