Single-cell analyses implicate ascites in remodeling the ecosystems of primary and metastatic tumors in ovarian cancer
- PMID: 37488416
- PMCID: PMC10447252
- DOI: 10.1038/s43018-023-00599-8
Single-cell analyses implicate ascites in remodeling the ecosystems of primary and metastatic tumors in ovarian cancer
Abstract
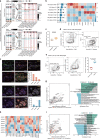
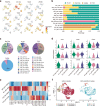
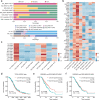
Ovarian cancer (OC) is an aggressive gynecological tumor usually diagnosed with widespread metastases and ascites. Here, we depicted a single-cell landscape of the OC ecosystem with five tumor-relevant sites, including omentum metastasis and malignant ascites. Our data reveal the potential roles of ascites-enriched memory T cells as a pool for tumor-infiltrating exhausted CD8+ T cells and T helper 1-like cells. Moreover, tumor-enriched macrophages exhibited a preference for monocyte-derived ontogeny, whereas macrophages in ascites were more of embryonic origin. Furthermore, we characterized MAIT and dendritic cells in malignant ascites, as well as two endothelial subsets in primary tumors as predictive biomarkers for platinum-based chemotherapy response. Taken together, our study provides a global view of the female malignant ascites ecosystem and offers valuable insights for its connection with tumor tissues and paves the way for potential markers of efficacy evaluation and therapy resistance in OC.
© 2023. The Author(s).
Conflict of interest statement
Z.H. is an employee of Analytical Biosciences. Z.Z. is a founder of Analytical Biosciences. All financial interests are unrelated to this study. The remaining authors declare no competing interests.
Figures















References
-
- Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019;69:280–304. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

