Identification and characterization of circRNAs in peri-implantation endometrium between Yorkshire and Erhualian pigs
- PMID: 37488487
- PMCID: PMC10364396
- DOI: 10.1186/s12864-023-09414-1
Identification and characterization of circRNAs in peri-implantation endometrium between Yorkshire and Erhualian pigs
Abstract
Background: One of the most critical periods for the loss of pig embryos is the 12th day of gestation when implantation begins. Recent studies have shown that non-coding RNAs (ncRNAs) play important regulatory roles during pregnancy. Circular RNAs (circRNAs) are a kind of ubiquitously expressed ncRNAs that can directly regulate the binding proteins or regulate the expression of target genes by adsorbing micro RNAs (miRNA).
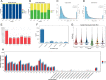
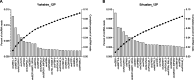
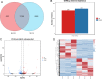
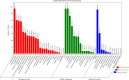
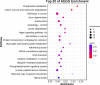
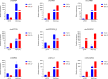
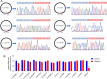
Results: We used the Illumina Novaseq6,000 technology to analyze the circRNA expression profile in the endometrium of three Erhualian (EH12) and three Yorkshire (YK12) pigs on day 12 of gestation. Overall, a total of 22,108 circRNAs were identified. Of these, 4051 circRNAs were specific to EH12 and 5889 circRNAs were specific to YK12, indicating a high level of breed specificity. Further analysis showed that there were 641 significant differentially expressed circRNAs (SDEcircRNAs) in EH12 compared with YK12 (FDR < 0.05). Functional enrichment of differential circRNA host genes revealed many pathways and genes associated with reproduction and regulation of embryo development. Network analysis of circRNA-miRNA interactions further supported the idea that circRNAs act as sponges for miRNAs to regulate gene expression. The prediction of differential circRNA binding proteins further explored the potential regulatory pathways of circRNAs. Analysis of SDEcircRNAs suggested a possible reason for the difference in embryo survival between the two breeds at the peri-implantation stage.
Conclusions: Together, these data suggest that circRNAs are abundantly expressed in the endometrium during the peri-implantation period in pigs and are important regulators of related genes. The results of this study will help to further understand the differences in molecular pathways between the two breeds during the critical implantation period of pregnancy, and will help to provide insight into the molecular mechanisms that contribute to the establishment of pregnancy and embryo loss in pigs.
Keywords: CircRNA; Embryo implantation; Endometrium; Erhualian; Peri-Implantation; Yorkshire.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
The authors declare no competing interests.
Figures



References
-
- Wähner M, Brüssow K. Biological potential of fecundity of sows. Biotechnol Anim Husb. 2009;25(5–6–1):523–33. doi: 10.2298/BAH0906523W. - DOI
-
- Argente MJ. Chap. 4 Major Components in Limiting Litter Size. In: 2018; 2018.
-
- Malopolska MM, Tuz R, Schwarz T, Ekanayake LD, D’Ambrosio J, Ahmadi B, Nowicki J, Tomaszewska E, Grzesiak M, Bartlewski PM. Correlates of reproductive tract anatomy and uterine histomorphometrics with fertility in swine. Theriogenology. 2021;165:44–51. doi: 10.1016/j.theriogenology.2021.02.007. - DOI - PubMed
-
- Ye TM, Pang RT, Leung CO, Chiu JF, Yeung WS. Two-dimensional liquid chromatography with tandem mass spectrometry-based proteomic characterization of endometrial luminal epithelial surface proteins responsible for embryo implantation. Fertil Steril. 2015;103(3):853–861e853. doi: 10.1016/j.fertnstert.2014.12.110. - DOI - PubMed
-
- Geisert RD, Johnson GA, Burghardt RC. Implantation and establishment of pregnancy in the Pig. Advances in anatomy, embryology, and cell biology 2015, 216:137–63. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

