The impact of SETBP1 mutations in neurological diseases and cancer
- PMID: 37489294
- PMCID: PMC11447826
- DOI: 10.1111/gtc.13057
The impact of SETBP1 mutations in neurological diseases and cancer
Erratum in
-
Correction to "The Impact of SETBP1 Mutations in Neurological Diseases and Cancer".Genes Cells. 2025 May;30(3):e70020. doi: 10.1111/gtc.70020. Genes Cells. 2025. PMID: 40254817 Free PMC article. No abstract available.
Abstract
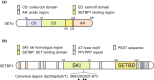
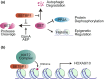
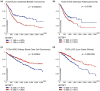
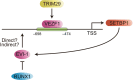
SE translocation (SET) is a cancer-promoting factor whose expression is upregulated in many cancers. High SET expression positively correlates with a poor cancer prognosis. SETBP1 (SET-binding protein 1/SEB/MRD29), identified as SET-binding protein, is the causative gene of Schinzel-Giedion syndrome, which is characterized by severe intellectual disability and a distorted facial appearance. Mutations in these genetic regions are also observed in some blood cancers, such as myelodysplastic syndromes, and are associated with a poor prognosis. However, the physiological role of SETBP1 and the molecular mechanisms by which the mutations lead to disease progression have not yet been fully elucidated. In this review, we will describe the current epidemiological data on SETBP1 mutations and shed light on the current knowledge about the SET-dependent and -independent functions of SETBP1.
Keywords: PP2A; SET; SETBP1; cancer; neurological disease.
© 2023 The Authors. Genes to Cells published by Molecular Biology Society of Japan and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
References
-
- Acuna‐Hidalgo, R. , Deriziotis, P. , Steehouwer, M. , Gilissen, C. , Graham, S. A. , van Dam, S. , Hoover‐Fong, J. , Telegrafi, A. B. , Destree, A. , Smigiel, R. , Lambie, L. A. , Kayserili, H. , Altunoglu, U. , Lapi, E. , Uzielli, M. L. , Aracena, M. , Nur, B. G. , Mihci, E. , Moreira, L. M. A. , … van Bon, B. W. (2017). Overlapping SETBP1 gain‐of‐function mutations in Schinzel‐Giedion syndrome and hematologic malignancies. PLoS Genetics, 13(3), e1006683. 10.1371/journal.pgen.1006683 - DOI - PMC - PubMed
-
- Adachi, Y. , Pavlakis, G. N. , & Copeland, T. D. (1994). Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. The Journal of Biological Chemistry, 269(3), 2258–2262. - PubMed
-
- Alsubaie, L. M. , Alsuwat, H. S. , Almandil, N. B. , AlSulaiman, A. , AbdulAzeez, S. , & Borgio, J. F. (2020). Risk Y‐haplotypes and pathogenic variants of Arab‐ancestry boys with autism by an exome‐wide association study. Molecular Biology Reports, 47(10), 7623–7632. 10.1007/s11033-020-05832-6 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

