Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms
- PMID: 37489388
- PMCID: PMC10366845
- DOI: 10.3390/proteomes11030022
Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms
Abstract
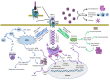
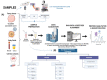
Recent advances in the field of proteomics have allowed extensive insights into the molecular regulations of the cell proteome. Specifically, this allows researchers to dissect a multitude of signaling arrays while targeting for the discovery of novel protein signatures. These approaches based on data mining are becoming increasingly powerful for identifying both potential disease mechanisms as well as indicators for disease progression and overall survival predictive and prognostic molecular markers for cancer. Furthermore, mass spectrometry (MS) integrations satisfy the ongoing demand for in-depth biomarker validation. For the purpose of this review, we will highlight the current developments based on MS sensitivity, to place quantitative proteomics into clinical settings and provide a perspective to integrate proteomics data for future applications in cancer precision medicine. We will also discuss malignancies associated with oncogenic viruses such as Acquire Immunodeficiency Syndrome (AIDS) and suggest novel mechanisms behind this phenomenon. Human Immunodeficiency Virus type-1 (HIV-1) proteins are known to be oncogenic per se, to induce oxidative and endoplasmic reticulum stresses, and to be released from the infected or expressing cells. HIV-1 proteins can act alone or in collaboration with other known oncoproteins, which cause the bulk of malignancies in people living with HIV-1 on ART.
Keywords: HIV-associated malignancies; biomarkers; cancer; co-morbidity; mass spectrometry; quantitative proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Al Shweiki M.R., Oeckl P., Steinacker P., Barschke P., Dorner-Ciossek C., Hengerer B., Schönfeldt-Lecuona C., Otto M. Proteomic Analysis Reveals a Biosignature of Decreased Synaptic Protein in Cerebrospinal Fluid of Major Depressive Disorder. Transl. Psychiatry. 2020;10:144. doi: 10.1038/s41398-020-0825-7. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

