Panaxynol alleviates colorectal cancer in a murine model via suppressing macrophages and inflammation
- PMID: 37489869
- PMCID: PMC10642997
- DOI: 10.1152/ajpgi.00119.2023
Panaxynol alleviates colorectal cancer in a murine model via suppressing macrophages and inflammation
Abstract
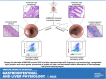
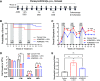
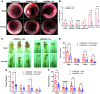
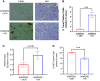
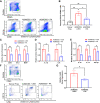
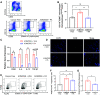
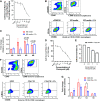
Currently available colorectal cancer (CRC) therapies have limited efficacy and severe adverse effects that may be overcome with the alternative use of natural compounds. We previously reported that panaxynol (PA), a bioactive component in American ginseng, possesses anticancer properties in vitro and suppresses murine colitis through its proapoptotic and anti-inflammatory properties. Because colitis is a predisposing factor of CRC and inflammation is a major driver of CRC, we sought to evaluate the therapeutic potential of PA in CRC. Azoxymethane-dextran sodium sulfate (AOM/DSS) mice (C57BL/6) were administered 2.5 mg/kg PA or vehicle 3 times/wk via oral gavage over 12 wk. PA improved clinical symptoms (P ≤ 0.05) and reduced tumorigenesis (P ≤ 0.05). This improvement may be reflective of PA's restorative effect on intestinal barrier function; PA upregulated the expression of essential tight junction and mucin genes (P ≤ 0.05) and increased the abundance of mucin-producing goblet cells (P ≤ 0.05). Given that macrophages play a substantial role in the pathogenesis of CRC and that we previously demonstrated that PA targets macrophages in colitis, we next assessed macrophages. We show that PA reduces the relative abundance of colonic macrophages within the lamina propria (P ≤ 0.05), and this was consistent with a reduction in the expression of important markers of macrophages and inflammation (P ≤ 0.05). We further confirmed PA's inhibitory effects on macrophages in vitro under CRC conditions (P ≤ 0.05). These results suggest that PA is a promising therapeutic compound to treat CRC and improve clinical symptoms given its ability to inhibit macrophages and modulate the inflammatory environment in the colon.NEW & NOTEWORTHY We report that panaxynol (PA) reduces colorectal cancer (CRC) by improving the colonic and tumor environment. Specifically, we demonstrate that PA improves crypt morphology, upregulates crucial tight junction and mucin genes, and promotes the abundance of mucin-producing goblet cells. Furthermore, PA reduces macrophages and associated inflammation, important drivers of CRC, in the colonic environment. This present study provides novel insights into the potential of PA as a therapeutic agent to ameliorate CRC tumorigenesis.
Keywords: AOM/DSS model; American ginseng; falcarinol; macrophages; natural compounds.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures

Comment in
-
Exploring the use of panaxynol from American ginseng to combat intestinal inflammation and colon cancer.Am J Physiol Gastrointest Liver Physiol. 2024 Aug 1;327(2):G120-G122. doi: 10.1152/ajpgi.00135.2024. Epub 2024 Jun 25. Am J Physiol Gastrointest Liver Physiol. 2024. PMID: 38915278 No abstract available.
References
-
- National Cancer Institute. SEER Stat Fact Sheets: Colon and Rectum Cancer (Online). National Cancer Institute, 2011. https://seer.cancer.gov/statfacts/html/colorect.html.
-
- Hale KE. Toxicities of chemotherapy. In: Principles of Critical Care (4th ed.), edited by Hall JB, Schmidt GA, Kress JP.. New York: McGraw-Hill Education, chapt. 95, 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

