Establishment of a CPER reverse genetics system for Powassan virus defines attenuating NS1 glycosylation sites and an infectious NS1-GFP11 reporter virus
- PMID: 37489888
- PMCID: PMC10470542
- DOI: 10.1128/mbio.01388-23
Establishment of a CPER reverse genetics system for Powassan virus defines attenuating NS1 glycosylation sites and an infectious NS1-GFP11 reporter virus
Abstract
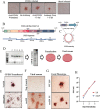
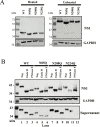
Powassan virus (POWV) is an emerging tick-borne Flavivirus that causes lethal encephalitis and long-term neurologic damage. Currently, there are no POWV therapeutics, licensed vaccines, or reverse genetics systems for producing infectious POWVs from recombinant DNA. Using a circular polymerase extension reaction (CPER), we generated recombinant LI9 (recLI9) POWVs with attenuating NS1 protein mutations and a recLI9-split-eGFP reporter virus. NS1 proteins are highly conserved glycoproteins that regulate replication, spread, and neurovirulence. POWV NS1 contains three putative N-linked glycosylation sites that we modified individually in infectious recLI9 mutants (N85Q, N208Q, and N224Q). NS1 glycosylation site mutations reduced replication kinetics and were attenuated, with 1-2 log decreases in titer. Severely attenuated recLI9-N224Q exhibited a 2- to 3-day delay in focal cell-to-cell spread and reduced NS1 secretion but was lethal when intracranially inoculated into suckling mice. However, footpad inoculation of recLI9-N224Q resulted in the survival of 80% of mice and demonstrated that NS1-N224Q mutations reduce POWV neuroinvasion in vivo. To monitor NS1 trafficking, we CPER fused a split GFP11-tag to the NS1 C-terminus and generated an infectious reporter virus, recLI9-NS1-GFP11. Cells infected with recLI9-NS1-GFP11 revealed NS1 trafficking in live cells and the novel formation of large NS1-lined intracellular vesicles. An infectious recLI9-NS1-GFP11 reporter virus permits real-time analysis of NS1 functions in POWV replication, assembly, and secretion and provides a platform for evaluating antiviral compounds. Collectively, our robust POWV reverse genetics system permits analysis of viral spread and neurovirulence determinants in vitro and in vivo and enables the rational genetic design of live attenuated POWV vaccines. IMPORTANCE Our findings newly establish a mechanism for genetically modifying Powassan viruses (POWVs), systematically defining pathogenic determinants and rationally designing live attenuated POWV vaccines. This initial study demonstrates that mutating POWV NS1 glycosylation sites attenuates POWV spread and neurovirulence in vitro and in vivo. Our findings validate a robust circular polymerase extension reaction approach as a mechanism for developing, and evaluating, attenuated genetically modified POWVs. We further designed an infectious GFP-tagged reporter POWV that permits us to monitor secretory trafficking of POWV in live cells, which can be applied to screen potential POWV replication inhibitors. This robust system for modifying POWVs provides the ability to define attenuating POWV mutations and create genetically attenuated recPOWV vaccines.
Keywords: NS1 glycosylation mutants; Powassan virus; attenuation; cell-to-cell spread; circular polymerase extension reaction; flavivirus; reverse genetics; split-GFP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Age-dependent Powassan virus lethality is linked to glial cell activation and divergent neuroinflammatory cytokine responses in a murine model.J Virol. 2024 Aug 20;98(8):e0056024. doi: 10.1128/jvi.00560-24. Epub 2024 Aug 1. J Virol. 2024. PMID: 39087762 Free PMC article.
-
Pathogenicity and virulence of Powassan virus.Virulence. 2025 Dec;16(1):2523887. doi: 10.1080/21505594.2025.2523887. Epub 2025 Jun 26. Virulence. 2025. PMID: 40545598 Free PMC article. Review.
-
Passage-attenuated Powassan virus LI9P protects mice from lethal LI9 challenge and links envelope residue D308 to neurovirulence.mBio. 2025 Apr 9;16(4):e0006525. doi: 10.1128/mbio.00065-25. Epub 2025 Feb 25. mBio. 2025. PMID: 39998203 Free PMC article.
-
Powassan Viruses Spread Cell to Cell during Direct Isolation from Ixodes Ticks and Persistently Infect Human Brain Endothelial Cells and Pericytes.J Virol. 2022 Jan 12;96(1):e0168221. doi: 10.1128/JVI.01682-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643436 Free PMC article.
-
Powassan Virus: An Emerging Arbovirus of Public Health Concern in North America.Vector Borne Zoonotic Dis. 2017 Jul;17(7):453-462. doi: 10.1089/vbz.2017.2110. Epub 2017 May 12. Vector Borne Zoonotic Dis. 2017. PMID: 28498740 Free PMC article. Review.
Cited by
-
Age-dependent Powassan virus lethality is linked to glial cell activation and divergent neuroinflammatory cytokine responses in a murine model.J Virol. 2024 Aug 20;98(8):e0056024. doi: 10.1128/jvi.00560-24. Epub 2024 Aug 1. J Virol. 2024. PMID: 39087762 Free PMC article.
-
Pathogenicity and virulence of Powassan virus.Virulence. 2025 Dec;16(1):2523887. doi: 10.1080/21505594.2025.2523887. Epub 2025 Jun 26. Virulence. 2025. PMID: 40545598 Free PMC article. Review.
-
A rapid and versatile reverse genetics approach for generating recombinant positive-strand RNA viruses that use IRES-mediated translation.J Virol. 2024 Mar 19;98(3):e0163823. doi: 10.1128/jvi.01638-23. Epub 2024 Feb 14. J Virol. 2024. PMID: 38353536 Free PMC article.
-
Passage-attenuated Powassan virus LI9P protects mice from lethal LI9 challenge and links envelope residue D308 to neurovirulence.mBio. 2025 Apr 9;16(4):e0006525. doi: 10.1128/mbio.00065-25. Epub 2025 Feb 25. mBio. 2025. PMID: 39998203 Free PMC article.
-
Reverse genetics rescue of sylvatic dengue viruses.J Virol. 2025 Jul 22;99(7):e0045025. doi: 10.1128/jvi.00450-25. Epub 2025 Jun 4. J Virol. 2025. PMID: 40464564 Free PMC article.
References
-
- Ebel GD, Kramer LD. 2004. Short report: duration of tick attachment required for transmission of Powassan virus by deer ticks. Am J Trop Med Hyg 71:268–271. - PubMed
Publication types
MeSH terms
Grants and funding
- R01AI12901005/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- RO1AI027044/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R21AI15237201/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R21AI13173902/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- T32AI007539/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical

