Loss of Mtm1 causes cholestatic liver disease in a model of X-linked myotubular myopathy
- PMID: 37490339
- PMCID: PMC10503795
- DOI: 10.1172/JCI166275
Loss of Mtm1 causes cholestatic liver disease in a model of X-linked myotubular myopathy
Abstract
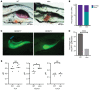
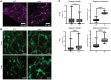
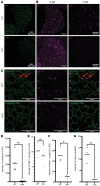
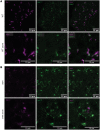
X-linked myotubular myopathy (XLMTM) is a fatal congenital disorder caused by mutations in the MTM1 gene. Currently, there are no approved treatments, although AAV8-mediated gene transfer therapy has shown promise in animal models and preliminarily in patients. However, 4 patients with XLMTM treated with gene therapy have died from progressive liver failure, and hepatobiliary disease has now been recognized more broadly in association with XLMTM. In an attempt to understand whether loss of MTM1 itself is associated with liver pathology, we have characterized what we believe to be a novel liver phenotype in a zebrafish model of this disease. Specifically, we found that loss-of-function mutations in mtm1 led to severe liver abnormalities including impaired bile flux, structural abnormalities of the bile canaliculus, and improper endosome-mediated trafficking of canalicular transporters. Using a reporter-tagged Mtm1 zebrafish line, we established localization of Mtm1 in the liver in association with Rab11, a marker of recycling endosomes, and canalicular transport proteins and demonstrated that hepatocyte-specific reexpression of Mtm1 could rescue the cholestatic phenotype. Last, we completed a targeted chemical screen and found that Dynasore, a dynamin-2 inhibitor, was able to partially restore bile flow and transporter localization to the canalicular membrane. In summary, we demonstrate, for the first time to our knowledge, liver abnormalities that were directly caused by MTM1 mutation in a preclinical model, thus establishing the critical framework for better understanding and comprehensive treatment of the human disease.
Keywords: Hepatology; Monogenic diseases; Muscle Biology.
Figures




References
-
- Pierson CR. Gene therapy strategies for X-linked myotubular myopathy. Expert Opin Orphan Drugs. 2018;6(3):193–202. doi: 10.1080/21678707.2018.1443807. - DOI

