Baseline Mutations and ctDNA Dynamics as Prognostic and Predictive Factors in ER-Positive/HER2-Negative Metastatic Breast Cancer Patients
- PMID: 37490393
- PMCID: PMC10570672
- DOI: 10.1158/1078-0432.CCR-23-0956
Baseline Mutations and ctDNA Dynamics as Prognostic and Predictive Factors in ER-Positive/HER2-Negative Metastatic Breast Cancer Patients
Abstract
Purpose: Prognostic and predictive biomarkers to cyclin-dependent kinases 4 and 6 inhibitors are lacking. Circulating tumor DNA (ctDNA) can be used to profile these patients and dynamic changes in ctDNA could be an early predictor of treatment efficacy. Here, we conducted plasma ctDNA profiling in patients from the PEARL trial comparing palbociclib+fulvestrant versus capecitabine to investigate associations between baseline genomic landscape and on-treatment ctDNA dynamics with treatment efficacy.
Experimental design: Correlative blood samples were collected at baseline [cycle 1-day 1 (C1D1)] and prior to treatment [cycle 1-day 15 (C1D15)]. Plasma ctDNA was sequenced with a custom error-corrected capture panel, with both univariate and multivariate Cox models used for treatment efficacy associations. A prespecified methodology measuring ctDNA changes in clonal mutations between C1D1 and C1D15 was used for the on-treatment ctDNA dynamic model.
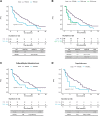
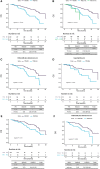
Results: 201 patients were profiled at baseline, with ctDNA detection associated with worse progression-free survival (PFS)/overall survival (OS). Detectable TP53 mutation showed worse PFS and OS in both treatment arms, even after restricting population to baseline ctDNA detection. ESR1 mutations were associated with worse OS overall, which was lost when restricting population to baseline ctDNA detection. PIK3CA mutations confer worse OS only to patients on the palbociclib+fulvestrant treatment arm. ctDNA dynamics analysis (n = 120) showed higher ctDNA suppression in the capecitabine arm. Patients without ctDNA suppression showed worse PFS in both treatment arms.
Conclusions: We show impaired survival irrespective of endocrine or chemotherapy-based treatments for patients with hormone receptor-positive/HER2-negative metastatic breast cancer harboring plasma TP53 mutations. Early ctDNA suppression may provide treatment efficacy predictions. Further validation to fully demonstrate clinical utility of ctDNA dynamics is warranted.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Finn RS, Martin M, Rugo HS, Jones S, Im S-A, Gelmon K, et al. . Palbociclib and letrozole in advanced breast cancer. N Engl J Med 2016;375:1925–36. - PubMed
-
- Hortobagyi GN, Stemmer SM, Burris HA, Yap Y-S, Sonke GS, Paluch-Shimon S, et al. . Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N Engl J Med 2016;375:1738–48. - PubMed
-
- Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. . MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J Clin Oncol 2017;35:3638–46. - PubMed
-
- Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, et al. . Palbociclib in hormone receptor–positive advanced breast cancer. N Engl J Med 2015;373:209–19. - PubMed
-
- Slamon DJ, Neven P, Chia S, Fasching PA, De Laurentiis M, Im S-A, et al. . Phase III randomized study of ribociclib and fulvestrant in hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: MONALEESA-3. J Clin Oncol 2018;36:2465–72. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

