The transcriptional regulators of virulence for Pseudomonas aeruginosa: Therapeutic opportunity and preventive potential of its clinical infections
- PMID: 37492705
- PMCID: PMC10363592
- DOI: 10.1016/j.gendis.2022.09.009
The transcriptional regulators of virulence for Pseudomonas aeruginosa: Therapeutic opportunity and preventive potential of its clinical infections
Abstract
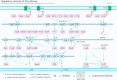
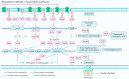
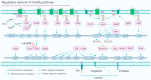
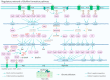
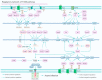
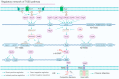
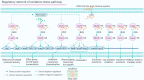
In Pseudomonas aeruginosa (P. aeruginosa), transcription factors (TFs) are important mediators in the genetic regulation of adaptability and pathogenicity to respond to multiple environmental stresses and host defences. The P. aeruginosa genome harbours 371 putative TFs; of these, about 70 have been shown to regulate virulence-associated phenotypes by binding to the promoters of their target genes. Over the past three decades, several techniques have been applied to identify TF binding sites on the P. aeruginosa genome, and an atlas of TF binding patterns has been mapped. The virulence-associated regulons of TFs show complex crosstalk in P. aeruginosa's regulatory network. In this review, we summarise the recent literature on TF regulatory networks involved in the quorum-sensing system, biofilm formation, pyocyanin synthesis, motility, the type III secretion system, the type VI secretion system, and oxidative stress responses. We discuss future perspectives that could provide insights and targets for preventing clinical infections caused by P. aeruginosa based on the global regulatory network of transcriptional regulators.
Keywords: Crosstalk; Pseudomonas aeruginosa; Regulatory network; Transcriptional regulators; Virulence.
© 2022 The Authors. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Figures
Similar articles
-
Virulence-related regulatory network of Pseudomonas syringae.Comput Struct Biotechnol J. 2022 Nov 9;20:6259-6270. doi: 10.1016/j.csbj.2022.11.011. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420163 Free PMC article. Review.
-
RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1.J Bacteriol. 2018 Jul 25;200(16):e00205-18. doi: 10.1128/JB.00205-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29760208 Free PMC article.
-
An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa.Nat Commun. 2019 Jul 3;10(1):2931. doi: 10.1038/s41467-019-10778-w. Nat Commun. 2019. PMID: 31270321 Free PMC article.
-
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article.
-
Novel therapeutic strategies for treating Pseudomonas aeruginosa infection.Expert Opin Drug Discov. 2020 Dec;15(12):1403-1423. doi: 10.1080/17460441.2020.1803274. Epub 2020 Sep 3. Expert Opin Drug Discov. 2020. PMID: 32880507 Review.
Cited by
-
CzcR-dependent reduction of catalase gene expression and induction of catalase activity in Pseudomonas aeruginosa during zinc excess.BMC Microbiol. 2024 Nov 29;24(1):509. doi: 10.1186/s12866-024-03671-0. BMC Microbiol. 2024. PMID: 39614155 Free PMC article.
-
An antibiotic-responsive regulator orchestrates chronic-to-acute virulence switch in Pseudomonas aeruginosa.Nucleic Acids Res. 2025 May 22;53(10):gkaf471. doi: 10.1093/nar/gkaf471. Nucleic Acids Res. 2025. PMID: 40464689 Free PMC article.
-
Naringenin as a Potent Natural Biofilm Inhibitor of Pseudomonas aeruginosa in Diabetic Foot Ulcers Through lasR Competitive Inhibition.Curr Microbiol. 2025 May 24;82(7):305. doi: 10.1007/s00284-025-04283-1. Curr Microbiol. 2025. PMID: 40413369
-
Whole-Genome Sequencing of Resistance, Virulence and Regulation Genes in Extremely Resistant Strains of Pseudomonas aeruginosa.Med Sci (Basel). 2025 Jan 3;13(1):6. doi: 10.3390/medsci13010006. Med Sci (Basel). 2025. PMID: 39846701 Free PMC article.
-
Structure and function of bacterial transcription regulators of the SorC family.Transcription. 2024 Jun-Oct;15(3-5):139-160. doi: 10.1080/21541264.2024.2387895. Epub 2024 Sep 3. Transcription. 2024. PMID: 39223991 Review.
References
-
- Deretic V., Schurr M.J., Yu H. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995;3(9):351–356. - PubMed
-
- Impey R.E., Panjikar S., Hall C.J., et al. Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation. FEBS J. 2020;287(2):386–400. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

