PATHWAYS ASSOCIATED WITH POSITIVE SEPSIS SURVIVAL OUTCOMES IN AFRICAN AMERICAN/BLACK AND NON-HISPANIC WHITE PATIENTS WITH URINARY TRACT INFECTION
- PMID: 37493584
- PMCID: PMC10527228
- DOI: 10.1097/SHK.0000000000002176
PATHWAYS ASSOCIATED WITH POSITIVE SEPSIS SURVIVAL OUTCOMES IN AFRICAN AMERICAN/BLACK AND NON-HISPANIC WHITE PATIENTS WITH URINARY TRACT INFECTION
Abstract
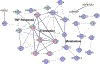
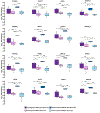
Urinary tract infections (UTIs) are a common cause of sepsis worldwide. Annually, more than 60,000 US deaths can be attributed to sepsis secondary to UTIs, and African American/Black adults have higher incidence and case-fatality rates than non-Hispanic White adults. Molecular-level factors that may help partially explain differences in sepsis survival outcomes between African American/Black and Non-Hispanic White adults are not clear. In this study, patient samples (N = 166) from the Protocolized Care for Early Septic Shock cohort were analyzed using discovery-based plasma proteomics. Patients had sepsis secondary to UTIs and were stratified according to self-identified racial background and sepsis survival outcomes. Proteomics results suggest patient heterogeneity across mechanisms driving survival from sepsis secondary to UTIs. Differentially expressed proteins (n = 122, false discovery rate-adjusted P < 0.05) in Non-Hispanic White sepsis survivors were primarily in immune system pathways, while differentially expressed proteins (n = 47, false discovery rate-adjusted P < 0.05) in African American/Black patients were mostly in metabolic pathways. However, in all patients, regardless of racial background, there were 16 differentially expressed proteins in sepsis survivors involved in translation initiation and shutdown pathways. These pathways are potential targets for prognostic intervention. Overall, this study provides information about molecular factors that may help explain disparities in sepsis survival outcomes among African American/Black and Non-Hispanic White patients with primary UTIs.
Copyright © 2023 by the Shock Society.
Conflict of interest statement
CONFLICTS OF INTEREST
Vanderbilt University and the University of Pittsburgh have jointly filed for patent protection for a panel of biomarkers arising from related work.
JAK holds stock in and is currently a full-time employee of Spectral Medical, Toronto, ON, and discloses consulting fees paid by Astute Medical, a BioMerieux company. Other authors declare no competing interests.
Figures
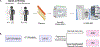


Similar articles
-
Proteomic changes associated with racial background and sepsis survival outcomes.Mol Omics. 2022 Dec 5;18(10):923-937. doi: 10.1039/d2mo00171c. Mol Omics. 2022. PMID: 36097965
-
Racial variation in the incidence, care, and outcomes of severe sepsis: analysis of population, patient, and hospital characteristics.Am J Respir Crit Care Med. 2008 Feb 1;177(3):279-84. doi: 10.1164/rccm.200703-480OC. Epub 2007 Nov 1. Am J Respir Crit Care Med. 2008. PMID: 17975201 Free PMC article.
-
Infection rate and acute organ dysfunction risk as explanations for racial differences in severe sepsis.JAMA. 2010 Jun 23;303(24):2495-503. doi: 10.1001/jama.2010.851. JAMA. 2010. PMID: 20571016 Free PMC article.
-
Racial Disparities in Sepsis-Related In-Hospital Mortality: Using a Broad Case Capture Method and Multivariate Controls for Clinical and Hospital Variables, 2004-2013.Crit Care Med. 2017 Dec;45(12):e1209-e1217. doi: 10.1097/CCM.0000000000002699. Crit Care Med. 2017. PMID: 28906287
-
Burden of present-on-admission infections and health care-associated infections, by race and ethnicity.Am J Infect Control. 2014 Dec;42(12):1296-302. doi: 10.1016/j.ajic.2014.08.019. Epub 2014 Nov 25. Am J Infect Control. 2014. PMID: 25465260 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

