Genetic Variants Associated With Hidradenitis Suppurativa
- PMID: 37494057
- PMCID: PMC10372759
- DOI: 10.1001/jamadermatol.2023.2217
Genetic Variants Associated With Hidradenitis Suppurativa
Abstract
Importance: Hidradenitis suppurativa (HS) is a common and severely morbid chronic inflammatory skin disease that is reported to be highly heritable. However, the genetic understanding of HS is insufficient, and limited genome-wide association studies (GWASs) have been performed for HS, which have not identified significant risk loci.
Objective: To identify genetic variants associated with HS and to shed light on the underlying genes and genetic mechanisms.
Design, setting, and participants: This genetic association study recruited 753 patients with HS in the HS Program for Research and Care Excellence (HS ProCARE) at the University of North Carolina Department of Dermatology from August 2018 to July 2021. A GWAS was performed for 720 patients (after quality control) with controls from the Add Health study and then meta-analyzed with 2 large biobanks, UK Biobank (247 cases) and FinnGen (673 cases). Variants at 3 loci were tested for replication in the BioVU biobank (290 cases). Data analysis was performed from September 2021 to December 2022.
Main outcomes and measures: Main outcome measures are loci identified, with association of P < 1 × 10-8 considered significant.
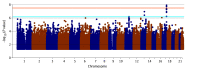
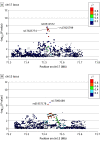
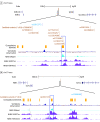
Results: A total of 753 patients were recruited, with 720 included in the analysis. Mean (SD) age at symptom onset was 20.3 (10.57) years and at enrollment was 35.3 (13.52) years; 360 (50.0%) patients were Black, and 575 (79.7%) were female. In a meta-analysis of the 4 studies, 2 HS-associated loci were identified and replicated, with lead variants rs10512572 (P = 2.3 × 10-11) and rs17090189 (P = 2.1 × 10-8) near the SOX9 and KLF5 genes, respectively. Variants at these loci are located in enhancer regulatory elements detected in skin tissue.
Conclusions and relevance: In this genetic association study, common variants associated with HS located near the SOX9 and KLF5 genes were associated with risk of HS. These or other nearby genes may be associated with genetic risk of disease and the development of clinical features, such as cysts, comedones, and inflammatory tunnels, that are unique to HS. New insights into disease pathogenesis related to these genes may help predict disease progression and novel treatment approaches in the future.
Conflict of interest statement
Figures
Comment in
-
Advances Toward the Clinical Translation of Hidradenitis Suppurativa Genetic Studies.JAMA Dermatol. 2023 Sep 1;159(9):913-915. doi: 10.1001/jamadermatol.2023.2205. JAMA Dermatol. 2023. PMID: 37494029 No abstract available.
References
-
- Alikhan A, Sayed C, Alavi A, et al. North American clinical management guidelines for hidradenitis suppurativa: a publication from the United States and Canadian Hidradenitis Suppurativa Foundations: part I: diagnosis, evaluation, and the use of complementary and procedural management. J Am Acad Dermatol. 2019;81(1):76-90. doi: 10.1016/j.jaad.2019.02.067 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

