Steroidogenic factor 1 (SF-1; Nr5a1) regulates the formation of the ovarian reserve
- PMID: 37494420
- PMCID: PMC10410717
- DOI: 10.1073/pnas.2220849120
Steroidogenic factor 1 (SF-1; Nr5a1) regulates the formation of the ovarian reserve
Abstract
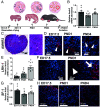
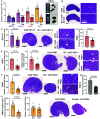
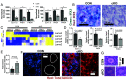
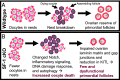
The ovarian follicle reserve, formed pre- or perinatally, comprises all oocytes for lifetime reproduction. Depletion of this reserve results in infertility. Steroidogenic factor 1 (SF-1; Nr5a1) and liver receptor homolog 1 (LRH-1; Nr5a2) are two orphan nuclear receptors that regulate adult endocrine function, but their role in follicle formation is unknown. We developed models of conditional depletion of SF-1 or LRH-1 from prenatal ovaries. Depletion of SF-1, but not LRH-1, resulted in dramatically smaller ovaries and fewer primordial follicles. This was mediated by increased oocyte death, resulting from increased ovarian inflammation and increased Notch signaling. Major dysregulated genes were Iroquois homeobox 3 and 5 and their downstream targets involved in the establishment of the ovarian laminin matrix and oocyte-granulosa cell gap junctions. Disruptions of these pathways resulted in follicles with impaired basement membrane formation and compromised oocyte-granulosa communication networks, believed to render them more prone to atresia. This study identifies SF-1 as a key regulator of the formation of the ovarian reserve.
Keywords: follicle assembly; orphan nuclear receptor; ovarian reserve; primordial follicle; steroidogenic factor 1.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Double knockout of steroidogenic factor 1 (NR5A1) and liver receptor homolog 1 (NR5A2) in the mouse ovary results in infertility due to disruption of follicle development and ovulation†.Biol Reprod. 2025 Jul 13;113(1):182-198. doi: 10.1093/biolre/ioaf079. Biol Reprod. 2025. PMID: 40184634
-
Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells.Biol Reprod. 2008 Dec;79(6):1074-83. doi: 10.1095/biolreprod.108.069435. Epub 2008 Aug 13. Biol Reprod. 2008. PMID: 18703422 Free PMC article.
-
Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary.Endocrinology. 2003 Aug;144(8):3598-610. doi: 10.1210/en.2002-0137. Endocrinology. 2003. PMID: 12865342
-
The Orphan Nuclear Receptors Steroidogenic Factor-1 and Liver Receptor Homolog-1: Structure, Regulation, and Essential Roles in Mammalian Reproduction.Physiol Rev. 2019 Apr 1;99(2):1249-1279. doi: 10.1152/physrev.00019.2018. Physiol Rev. 2019. PMID: 30810078 Review.
-
Orphan nuclear receptor function in the ovary.Front Biosci. 2007 May 1;12:3398-405. doi: 10.2741/2321. Front Biosci. 2007. PMID: 17485308 Review.
Cited by
-
Exploration of transcriptional regulation network between buffalo oocytes and granulosa cells and its impact on different diameter follicles.BMC Genomics. 2024 Oct 26;25(1):1004. doi: 10.1186/s12864-024-10912-z. BMC Genomics. 2024. PMID: 39462339 Free PMC article.
-
Single-cell sequencing reveals the transcriptional alternations of 17β-estradiol suppressing primordial follicle formation in neonatal mouse ovaries.Cell Prolif. 2024 Sep;57(9):e13713. doi: 10.1111/cpr.13713. Epub 2024 Jul 10. Cell Prolif. 2024. PMID: 38988058 Free PMC article.
-
Steroidogenic Factor-1 form and function: From phospholipids to physiology.Adv Biol Regul. 2024 Jan;91:100991. doi: 10.1016/j.jbior.2023.100991. Epub 2023 Sep 27. Adv Biol Regul. 2024. PMID: 37802761 Free PMC article. Review.
-
Role of epigenetic regulation in diminished ovarian reserve.J Assist Reprod Genet. 2025 Feb;42(2):389-403. doi: 10.1007/s10815-024-03301-8. Epub 2024 Dec 7. J Assist Reprod Genet. 2025. PMID: 39644448 Free PMC article. Review.
-
Phosphoinositide signalling in cell motility and adhesion.Nat Cell Biol. 2025 May;27(5):736-748. doi: 10.1038/s41556-025-01647-4. Epub 2025 Apr 1. Nat Cell Biol. 2025. PMID: 40169755 Review.
References
-
- Damario M. A., General aspects of fertility and infertility. Methods Mol. Biol. 1154, 3–23 (2014). - PubMed
-
- Vander Borght M., Wyns C., Fertility and infertility: Definition and epidemiology. Clin. Biochem. 62, 2–10 (2018). - PubMed
-
- Lew R., Natural history of ovarian function including assessment of ovarian reserve and premature ovarian failure. Best Pract. Res. Clin. Obstet. Gynaecol. 55, 2–13 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

