Cleavage of cFLIP restrains cell death during viral infection and tissue injury and favors tissue repair
- PMID: 37494451
- PMCID: PMC10371024
- DOI: 10.1126/sciadv.adg2829
Cleavage of cFLIP restrains cell death during viral infection and tissue injury and favors tissue repair
Abstract
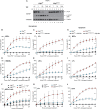
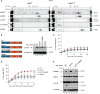
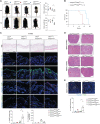
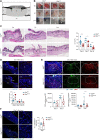
Cell death coordinates repair programs following pathogen attack and tissue injury. However, aberrant cell death can interfere with such programs and cause organ failure. Cellular FLICE-like inhibitory protein (cFLIP) is a crucial regulator of cell death and a substrate of Caspase-8. However, the physiological role of cFLIP cleavage by Caspase-8 remains elusive. Here, we found an essential role for cFLIP cleavage in restraining cell death in different pathophysiological scenarios. Mice expressing a cleavage-resistant cFLIP mutant, CflipD377A, exhibited increased sensitivity to severe acute respiratory syndrome coronavirus (SARS-CoV)-induced lethality, impaired skin wound healing, and increased tissue damage caused by Sharpin deficiency. In vitro, abrogation of cFLIP cleavage sensitizes cells to tumor necrosis factor(TNF)-induced necroptosis and apoptosis by favoring complex-II formation. Mechanistically, the cell death-sensitizing effect of the D377A mutation depends on glutamine-469. These results reveal a crucial role for cFLIP cleavage in controlling the amplitude of cell death responses occurring upon tissue stress to ensure the execution of repair programs.
Figures



References
-
- A. Annibaldi, P. Meier, Checkpoints in TNF-induced cell death: Implications in inflammation and cancer. Trends Mol. Med. 24, 49–65 (2018). - PubMed
-
- J. Zinngrebe, E. Rieser, L. Taraborrelli, N. Peltzer, T. Hartwig, H. Ren, I. Kovács, C. Endres, P. Draber, M. Darding, S. von Karstedt, J. Lemke, B. Dome, M. Bergmann, B. J. Ferguson, H. Walczak, LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689 (2016). - PMC - PubMed
-
- F. Balkwill, Tumour necrosis factor and cancer. Nat. Rev. Cancer 9, 361–371 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

