Parallel CRISPR-Cas9 screens identify mechanisms of PLIN2 and lipid droplet regulation
- PMID: 37494933
- PMCID: PMC10530302
- DOI: 10.1016/j.devcel.2023.07.001
Parallel CRISPR-Cas9 screens identify mechanisms of PLIN2 and lipid droplet regulation
Abstract
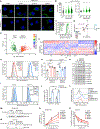
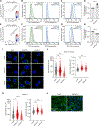
Despite the key roles of perilipin-2 (PLIN2) in governing lipid droplet (LD) metabolism, the mechanisms that regulate PLIN2 levels remain incompletely understood. Here, we leverage a set of genome-edited human PLIN2 reporter cell lines in a series of CRISPR-Cas9 loss-of-function screens, identifying genetic modifiers that influence PLIN2 expression and post-translational stability under different metabolic conditions and in different cell types. These regulators include canonical genes that control lipid metabolism as well as genes involved in ubiquitination, transcription, and mitochondrial function. We further demonstrate a role for the E3 ligase MARCH6 in regulating triacylglycerol biosynthesis, thereby influencing LD abundance and PLIN2 stability. Finally, our CRISPR screens and several published screens provide the foundation for CRISPRlipid (http://crisprlipid.org), an online data commons for lipid-related functional genomics data. Our study identifies mechanisms of PLIN2 and LD regulation and provides an extensive resource for the exploration of LD biology and lipid metabolism.
Keywords: CRISPR; ERAD; MARCH6; PLIN2; endoplasmic reticulum; genetic screen; lipid droplet; metabolism; perilipin; resource.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.A.O. is a member of the scientific advisory board for Vicinitas Therapeutics and has patent applications related to ferroptosis. M.K. is an inventor on a US patent related to CRISPRi and CRISPRa screening; serves on the scientific advisory boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience and Alector; and is a consultant to Modulo Bio and Recursion Therapeutics. F.F. and M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research. M.A.N. also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc. M.C.B. has outside interest in DEM Biopharma.
Figures