In-Depth Specificity Profiling of Endopeptidases Using Dedicated Mix-and-Split Synthetic Peptide Libraries and Mass Spectrometry
- PMID: 37495545
- PMCID: PMC10413326
- DOI: 10.1021/acs.analchem.3c01215
In-Depth Specificity Profiling of Endopeptidases Using Dedicated Mix-and-Split Synthetic Peptide Libraries and Mass Spectrometry
Abstract
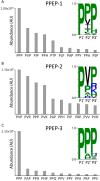
Proteases comprise the class of enzymes that catalyzes the hydrolysis of peptide bonds, thereby playing a pivotal role in many aspects of life. The amino acids surrounding the scissile bond determine the susceptibility toward protease-mediated hydrolysis. A detailed understanding of the cleavage specificity of a protease can lead to the identification of its endogenous substrates, while it is also essential for the design of inhibitors. Although many methods for protease activity and specificity profiling exist, none of these combine the advantages of combinatorial synthetic libraries, i.e., high diversity, equimolar concentration, custom design regarding peptide length, and randomization, with the sensitivity and detection power of mass spectrometry. Here, we developed such a method and applied it to study a group of bacterial metalloproteases that have the unique specificity to cleave between two prolines, i.e., Pro-Pro endopeptidases (PPEPs). We not only confirmed the prime-side specificity of PPEP-1 and PPEP-2, but also revealed some new unexpected peptide substrates. Moreover, we have characterized a new PPEP (PPEP-3) that has a prime-side specificity that is very different from that of the other two PPEPs. Importantly, the approach that we present in this study is generic and can be extended to investigate the specificity of other proteases.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

